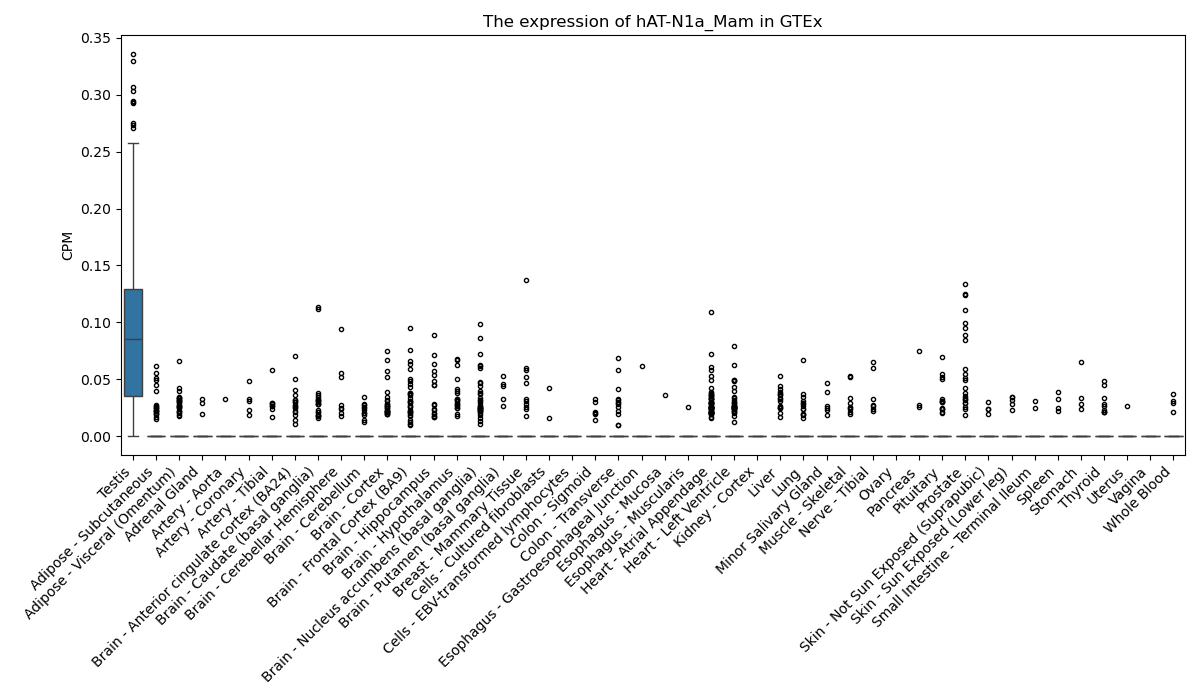
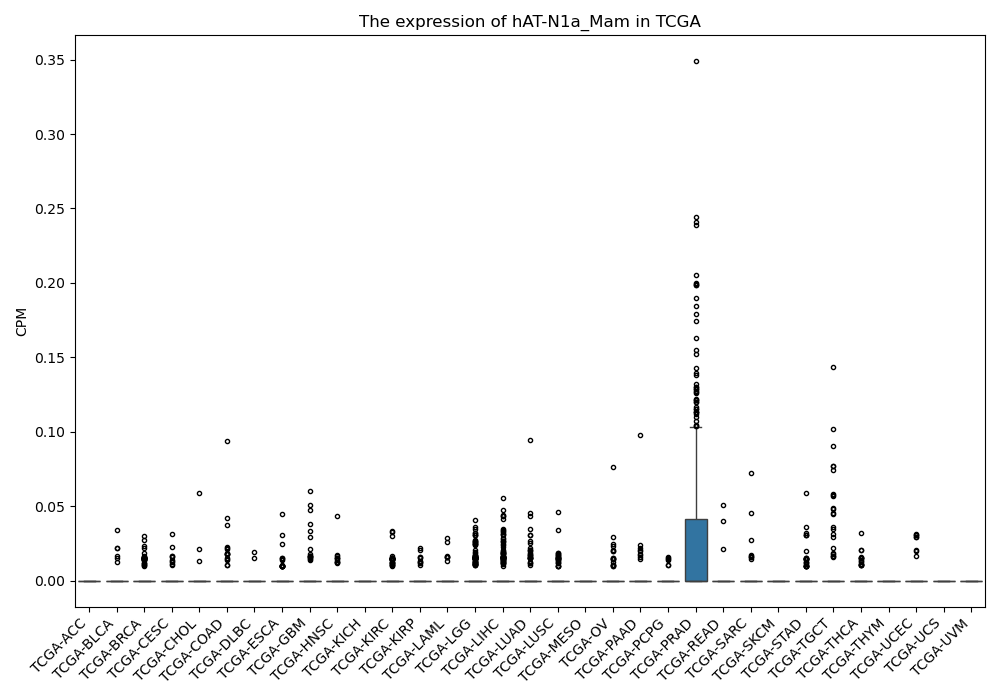
hAT-N1a_Mam
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001245 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 362 |
| Kimura value | 31.23 |
| Tau index | 1.0000 |
| Description | hAT-Tip100 DNA transposon, hAT-N1a_Mam subfamily |
| Comment | Incomplete at 5'end where the consensus runs in a simple repeat. Orientation may be the other way around. |
| Sequence |
GTGTGTGTGTGATGAGGATTAAACNGTACATGGTATCAGGGAAAACTGTCGACATTGATGCTGATTATAGATACTGGAGAACTAACAAAGTAAGAAGAGAAAAACTTAAAAATGCTTGGTTTGGCTGTGTAAAAGTAATACTAATTACAGTCATTTACGTGAACTGCTTACCATTTATGCCTAATTACCATGCAGCACAATAACGATTATGCATATCATTAAATATGTGTCAGATTAACTTTGTTGTAAGTTTTATTTGATTACAACACTAAACTCTGTTCACGTATGCTCAGTCTTCATAGTTTGTTGCTCATGTAGCATGGTCGTCTGCTCACAACTTTAAAAAATTAGAGGGAACATTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| hAT-N1a_Mam | Dfd | 216 | 222 | - | 12.20 | TTAATGA |
| hAT-N1a_Mam | Antp | 216 | 222 | - | 12.19 | TTAATGA |
| hAT-N1a_Mam | pdm3 | 141 | 148 | + | 12.15 | CTAATTAC |
| hAT-N1a_Mam | pdm3 | 181 | 188 | + | 12.15 | CTAATTAC |
| hAT-N1a_Mam | Dmbx1 | 12 | 21 | + | 12.11 | ATGAGGATTA |
| hAT-N1a_Mam | MYB62 | 179 | 187 | - | 12.02 | TAATTAGGC |
| hAT-N1a_Mam | Scr | 216 | 222 | - | 11.99 | TTAATGA |
| hAT-N1a_Mam | LHX9 | 141 | 147 | + | 11.98 | CTAATTA |
| hAT-N1a_Mam | LHX9 | 181 | 187 | + | 11.98 | CTAATTA |
| hAT-N1a_Mam | PRRX2 | 141 | 147 | + | 11.96 | CTAATTA |
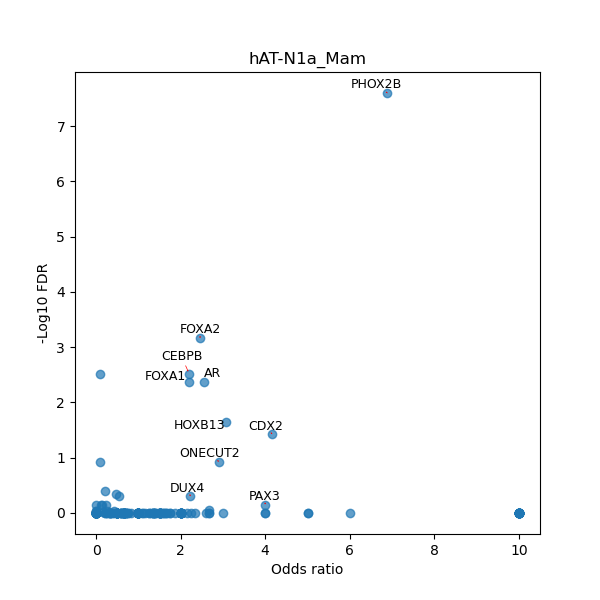
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.