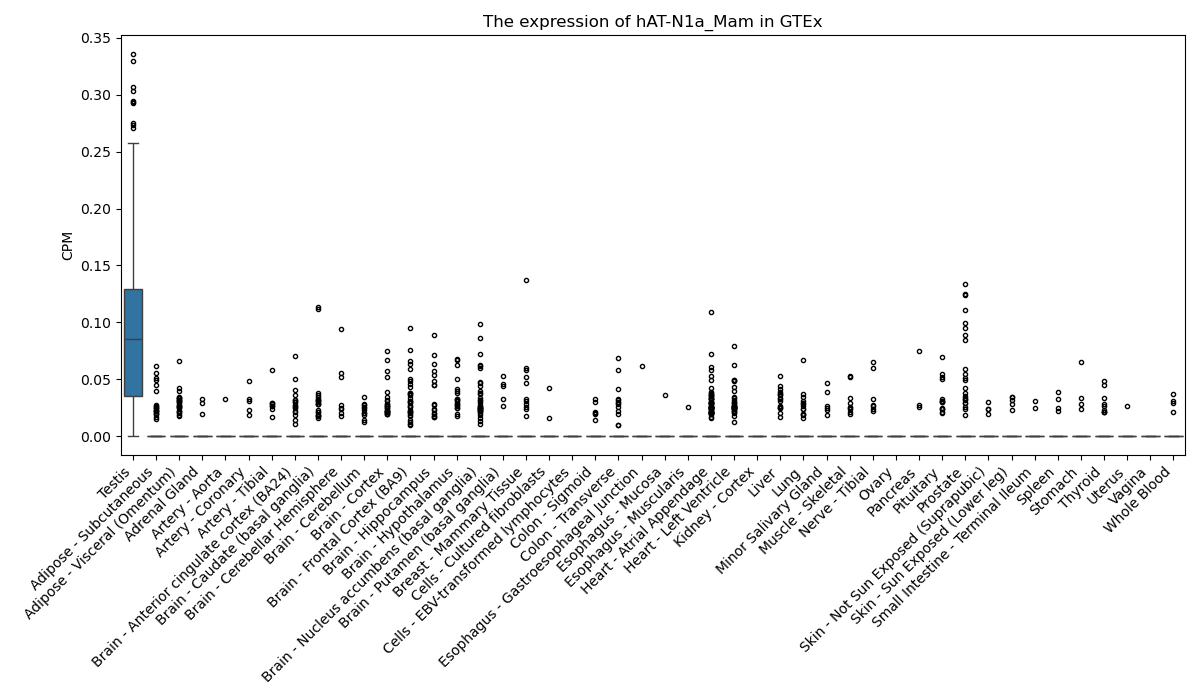
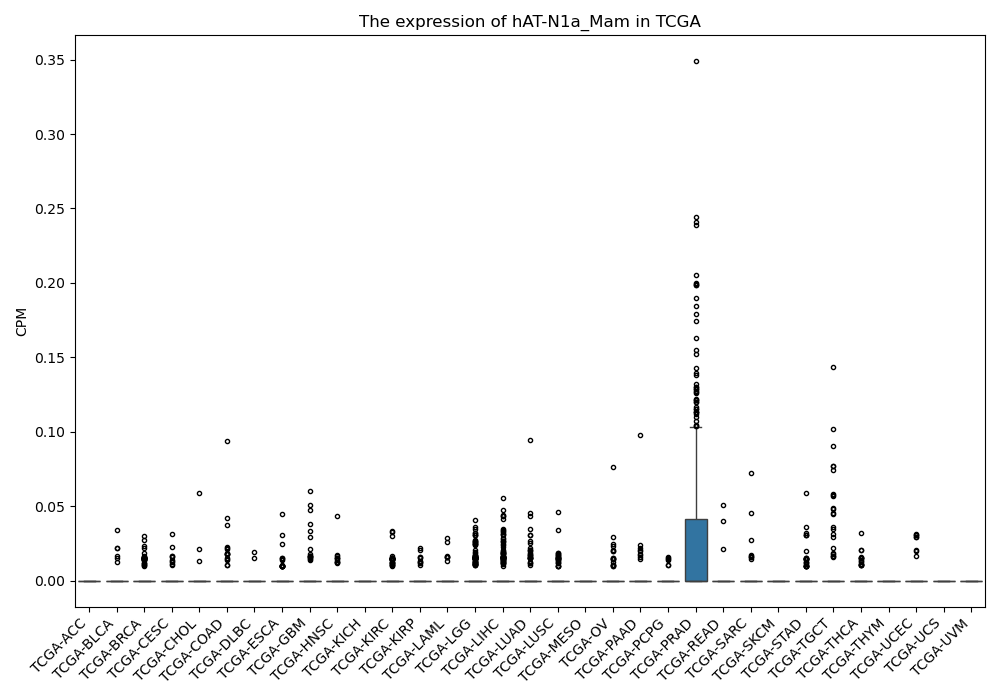
hAT-N1a_Mam
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001245 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 362 |
| Kimura value | 31.23 |
| Tau index | 1.0000 |
| Description | hAT-Tip100 DNA transposon, hAT-N1a_Mam subfamily |
| Comment | Incomplete at 5'end where the consensus runs in a simple repeat. Orientation may be the other way around. |
| Sequence |
GTGTGTGTGTGATGAGGATTAAACNGTACATGGTATCAGGGAAAACTGTCGACATTGATGCTGATTATAGATACTGGAGAACTAACAAAGTAAGAAGAGAAAAACTTAAAAATGCTTGGTTTGGCTGTGTAAAAGTAATACTAATTACAGTCATTTACGTGAACTGCTTACCATTTATGCCTAATTACCATGCAGCACAATAACGATTATGCATATCATTAAATATGTGTCAGATTAACTTTGTTGTAAGTTTTATTTGATTACAACACTAAACTCTGTTCACGTATGCTCAGTCTTCATAGTTTGTTGCTCATGTAGCATGGTCGTCTGCTCACAACTTTAAAAAATTAGAGGGAACATTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| hAT-N1a_Mam | HMRA1 | 195 | 201 | + | 12.91 | GCACAAT |
| hAT-N1a_Mam | GLYMA-07G038400 | 227 | 233 | - | 12.89 | CTGACAC |
| hAT-N1a_Mam | squamosa | 168 | 180 | - | 12.74 | GCATAAATGGTAA |
| hAT-N1a_Mam | ZHD10 | 140 | 147 | - | 12.70 | TAATTAGT |
| hAT-N1a_Mam | blmp-1 | 93 | 103 | + | 12.62 | AGAAGAGAAAA |
| hAT-N1a_Mam | ZHD5 | 141 | 148 | - | 12.45 | GTAATTAG |
| hAT-N1a_Mam | ZHD5 | 181 | 188 | - | 12.45 | GTAATTAG |
| hAT-N1a_Mam | PITX2 | 15 | 22 | - | 12.42 | TTAATCCT |
| hAT-N1a_Mam | POU3F1 | 207 | 216 | + | 12.25 | TTATGCATAT |
| hAT-N1a_Mam | KLF9 | 1 | 11 | - | 12.20 | CACACACACAC |
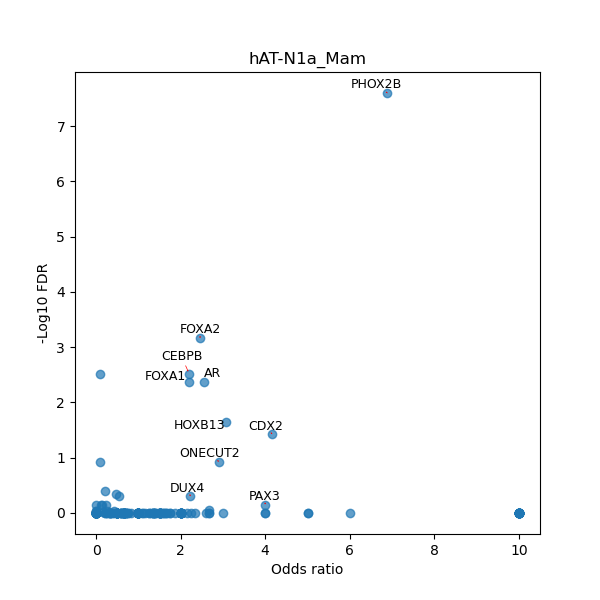
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.