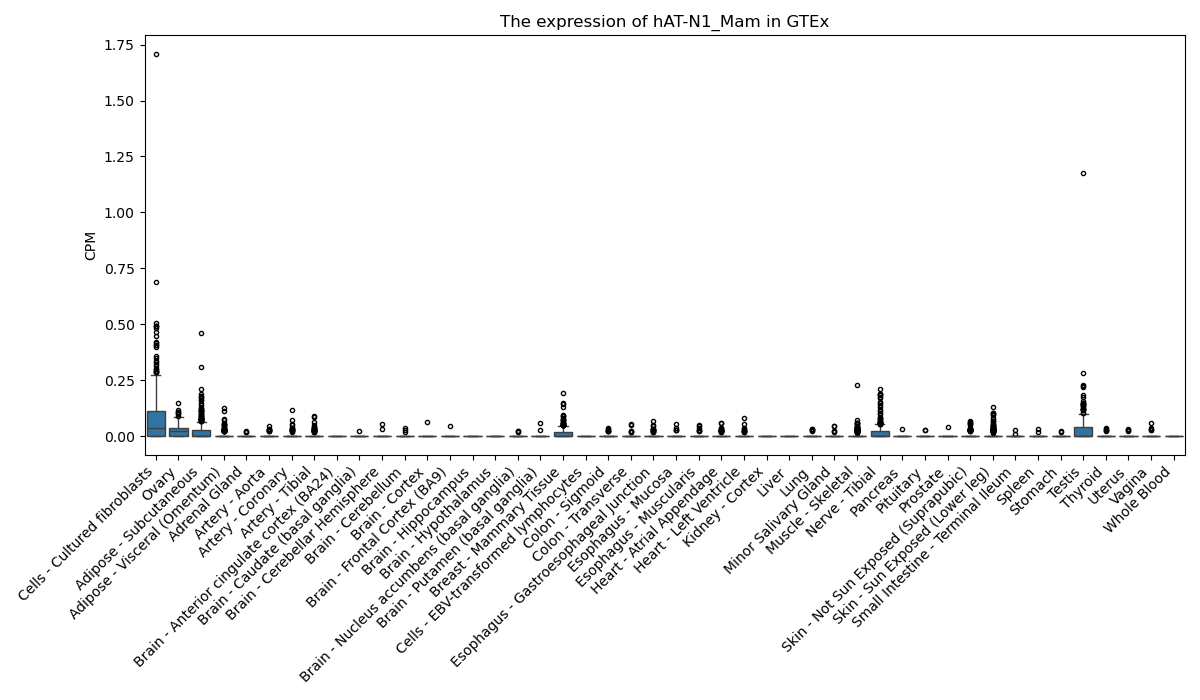
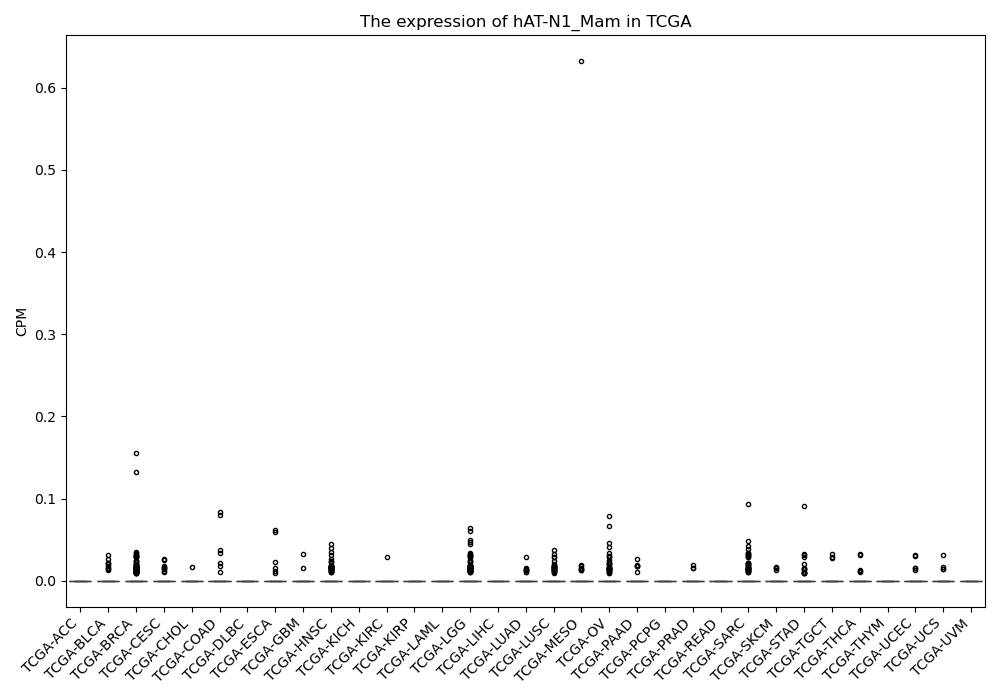
hAT-N1_Mam
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000978 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 340 |
| Kimura value | 32.76 |
| Tau index | 0.9868 |
| Description | Putative hAT-Tip10 DNA transposon, hAT-N1_Mam subfamily |
| Comment | hAT-N1_Mam is classified by 3' end similarities to hAT elements in Fugu and in the flatworm Schmidtea. The 5' end is uncertain, and no TSDs are apparent. |
| Sequence |
CTGGGGATCNGGGGNTCTTCAGAGGAGCCTCAGAGCAGCTTCCTGCTCTGCCCGGTTCCTCCCCCCCTTCCCCCCAGCTGACCCCAGAAATGCTTGGTTTGGCTGTGTAAAAGTAATACTAATTACAGTCATTTACGTGAACTGCTTACCATTTATGCCTAATTACCATGCAGCACAATAACGATTATGCATATCATTAAATATGTGTCAGATTAACTTTGTTGTAAGTTTTATTTGATTACAACACTAAACTCTGTTCACGTATGCTCAGTCTTCATAGTTTGTTGCTCACGTAGCATGGTCGTCTGCTCACAACTTTAAAAAATTAGAGGGAACATTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| hAT-N1_Mam | ceh-18 | 184 | 191 | - | 13.35 | TGCATAAT |
| hAT-N1_Mam | GRF6 | 206 | 212 | - | 13.30 | TCTGACA |
| hAT-N1_Mam | unc-86 | 185 | 192 | + | 13.03 | TTATGCAT |
| hAT-N1_Mam | GRF9 | 206 | 212 | + | 13.03 | TGTCAGA |
| hAT-N1_Mam | HMRA1 | 173 | 179 | + | 12.91 | GCACAAT |
| hAT-N1_Mam | GLYMA-07G038400 | 205 | 211 | - | 12.89 | CTGACAC |
| hAT-N1_Mam | Wt1 | 58 | 67 | + | 12.86 | CCTCCCCCCC |
| hAT-N1_Mam | squamosa | 146 | 158 | - | 12.74 | GCATAAATGGTAA |
| hAT-N1_Mam | ZHD10 | 118 | 125 | - | 12.70 | TAATTAGT |
| hAT-N1_Mam | usp | 77 | 85 | - | 12.54 | GGGGTCAGC |
TFBS enrichment in GRCh38
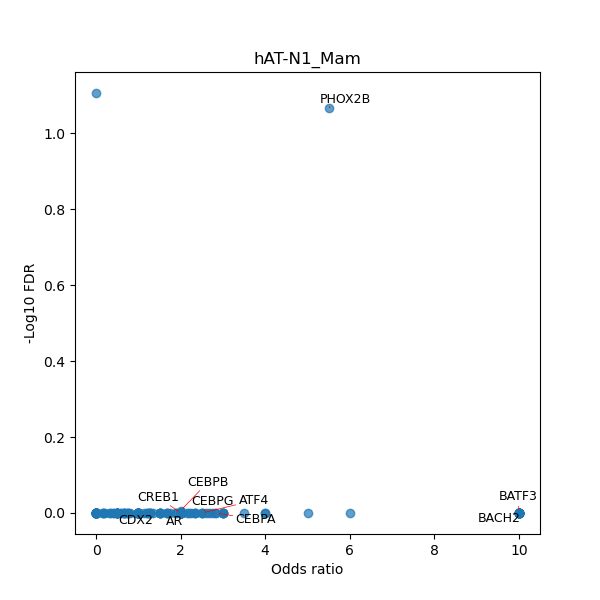
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.