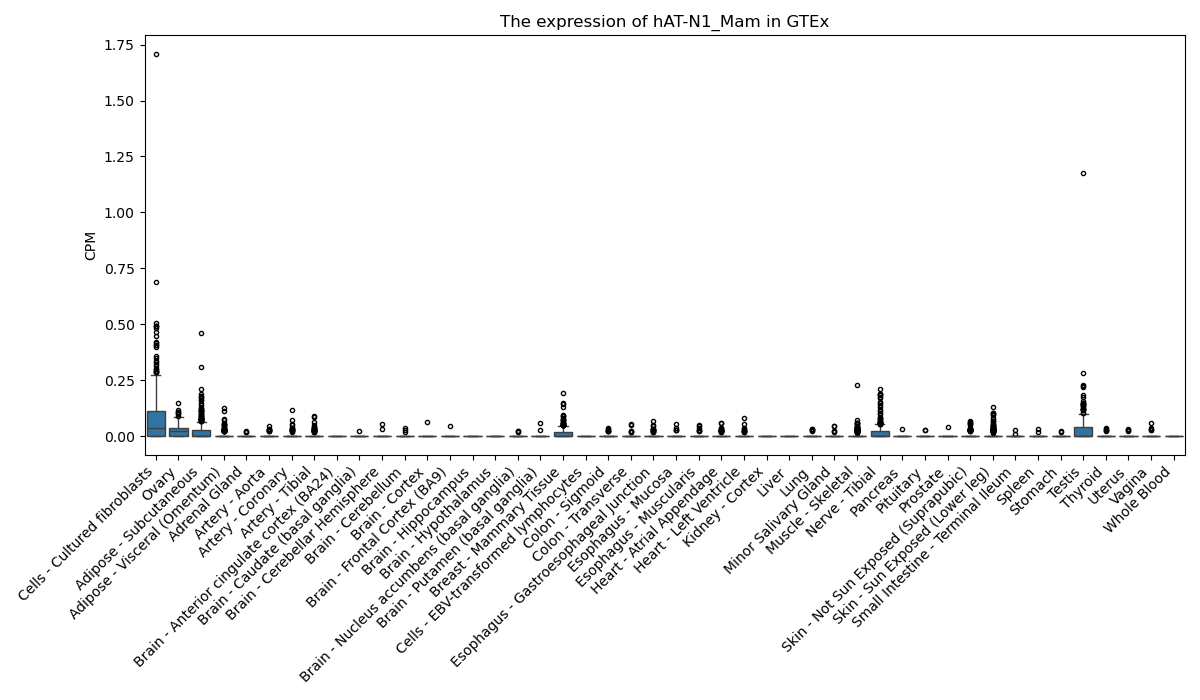
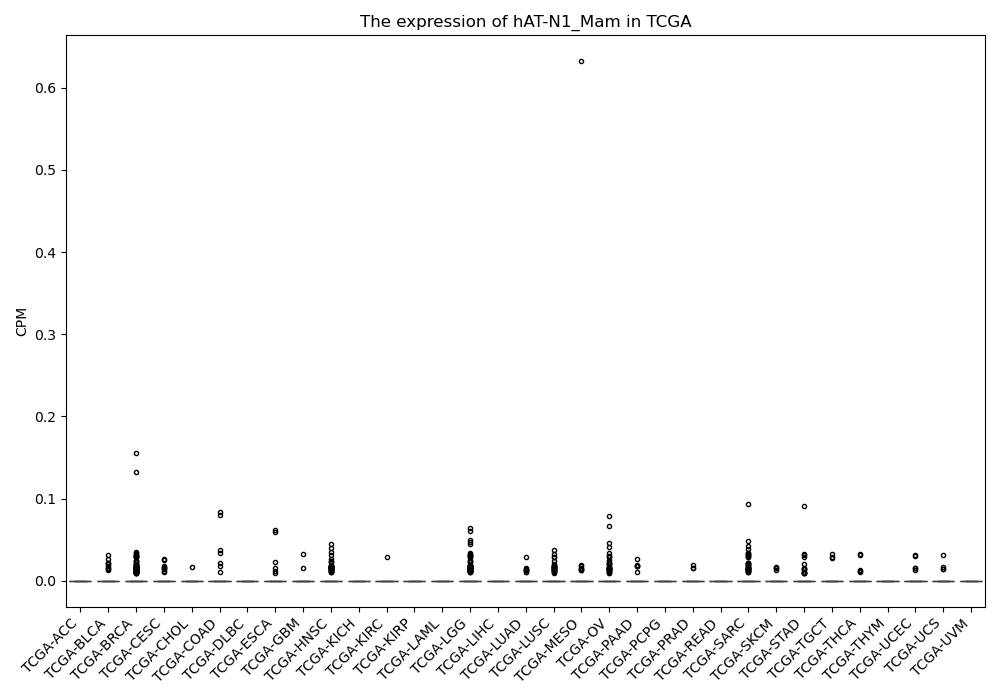
hAT-N1_Mam
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000978 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 340 |
| Kimura value | 32.76 |
| Tau index | 0.9868 |
| Description | Putative hAT-Tip10 DNA transposon, hAT-N1_Mam subfamily |
| Comment | hAT-N1_Mam is classified by 3' end similarities to hAT elements in Fugu and in the flatworm Schmidtea. The 5' end is uncertain, and no TSDs are apparent. |
| Sequence |
CTGGGGATCNGGGGNTCTTCAGAGGAGCCTCAGAGCAGCTTCCTGCTCTGCCCGGTTCCTCCCCCCCTTCCCCCCAGCTGACCCCAGAAATGCTTGGTTTGGCTGTGTAAAAGTAATACTAATTACAGTCATTTACGTGAACTGCTTACCATTTATGCCTAATTACCATGCAGCACAATAACGATTATGCATATCATTAAATATGTGTCAGATTAACTTTGTTGTAAGTTTTATTTGATTACAACACTAAACTCTGTTCACGTATGCTCAGTCTTCATAGTTTGTTGCTCACGTAGCATGGTCGTCTGCTCACAACTTTAAAAAATTAGAGGGAACATTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| hAT-N1_Mam | ERF::NHLH1 | 34 | 49 | + | 6.40 | AGCAGCTTCCTGCTCT |
| hAT-N1_Mam | ZIC5 | 69 | 83 | + | 5.72 | TCCCCCCAGCTGACC |
| hAT-N1_Mam | Atf6 | 286 | 298 | + | 4.72 | TGCTCACGTAGCA |
| hAT-N1_Mam | ZNF157 | 131 | 151 | - | 4.56 | TGGTAAGCAGTTCACGTAAAT |
| hAT-N1_Mam | EWSR1-FLI1 | 54 | 71 | - | 3.11 | GGAAGGGGGGGAGGAACC |
| hAT-N1_Mam | Fer1 | 71 | 83 | + | 2.92 | CCCCCAGCTGACC |
| hAT-N1_Mam | RARA | 68 | 84 | - | 2.61 | GGGTCAGCTGGGGGGAA |
| hAT-N1_Mam | EWSR1-FLI1 | 58 | 75 | - | -6.14 | GGGGGGAAGGGGGGGAGG |
| hAT-N1_Mam | EWSR1-FLI1 | 57 | 74 | - | -15.70 | GGGGGAAGGGGGGGAGGA |
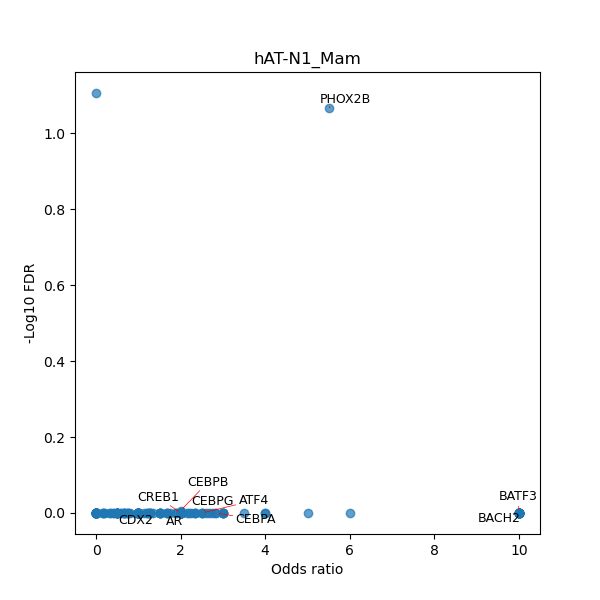
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.