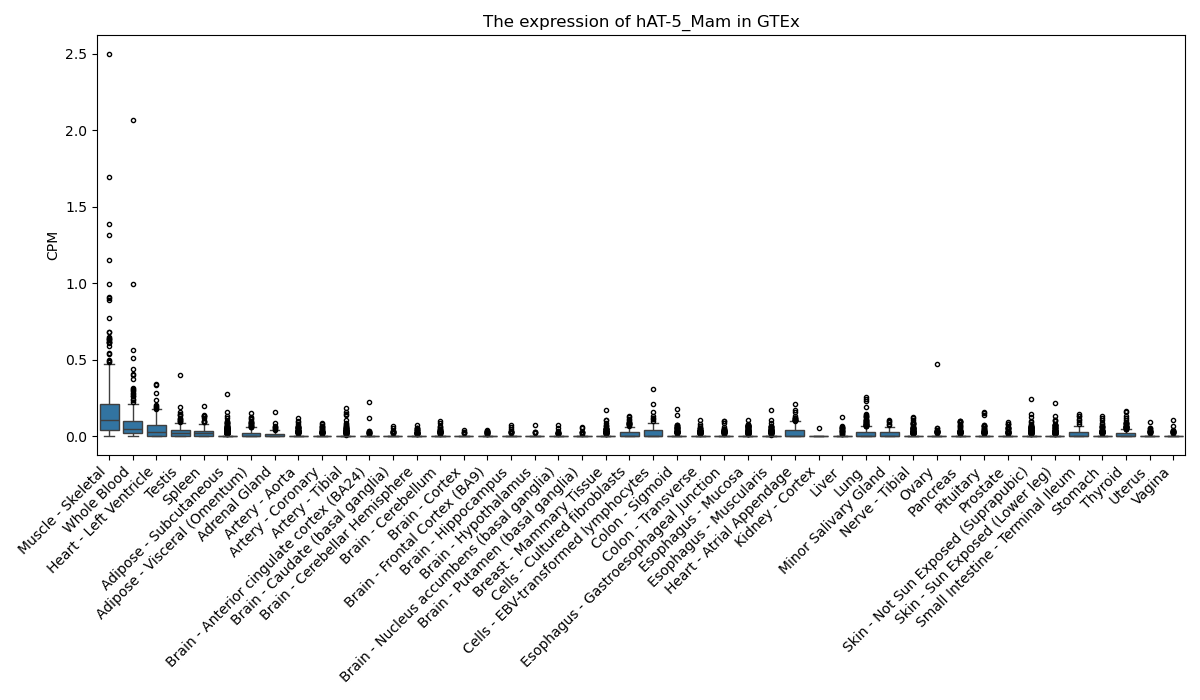
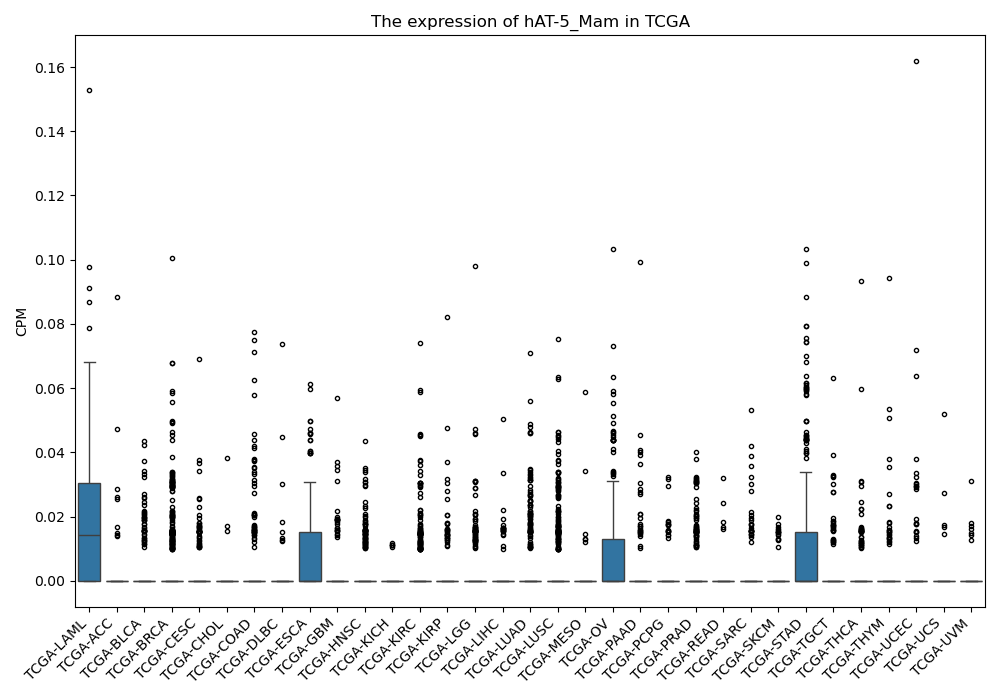
hAT-5_Mam
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001335 |
|---|---|
| TE superfamily | Tag1 |
| TE class | DNA |
| Species | Mammalia |
| Length | 3540 |
| Kimura value | 32.08 |
| Tau index | 0.9753 |
| Description | hAT-Tip100 DNA transposon, hAT-5_Mam subfamily |
| Comment | - |
| Sequence |
CAGGGTAGATAAAAATCNATNATTTTTNAAAAAAAAATTAAAAATCGGATTTTTTAATTTAATCGGATTTTNTANTTTTTNTNGTTAAGATGTTTTNTTAAGAAAAAGAAACCTATATAAAGATAGTTTTAATTTAAGATATGTTAAAGCTCAAAGATATCATCATGGAATAGGGATTATAACTTCTAATTATATANTATTTGAGACAATATATTCATGTAACCTTTTTAGAAAAGTTTTATAAATAGGTCACAACAGGCCACGGACTATATTAGGGGCCCAATCTTATGGGGTTCCCAGAGGCTCCTCTATAGATTANTNAGAAAGATTAAAGAAAACCACTCTACCCAATGGGACTCAGTGCTCAGTCGATCTAGAAGATCCCATTAAGCATCTCTGTGATGCTTAGTTTCAGTTCTCAAACTGTGGATTTGTGTCTCCAGAGATAACATCCTTGTTAACAGCAGTATATGGAGATGAGAGATAACAGACCTGATCAGCCCTATTGTCTCTCTGCAAGTTTGTGTACACAGAGTCAAGCCCTTACCTCTCTCTAAAAGTGCAAAGTTTCAAGAAGTTAAATGAATAGAGGATTGTTGGGGGTGGAGTAGATCTGGGCAAGGAGGAGGAGTCTGGAGATAAATGCAAGGAGGGAGGGGCAGTAGAAACAAAAGAGAAACATTTTCGAGCAGAATATTCCAGAAGTCTTGAGGGATAAGTTTCTGAGTATAGCTTCCATTGATTTGAGATCTAGTGAGGGAATGGTATGGTAGAAGGGAAAACCTATCATGGCAGCAGGCCGTAAAAGAGACCCGTTTGGGAATATTTTAATGAAGTTCCTCTNCCTGTGGGTAAGATAGGCATGCGTGCAAAATGCAAACAGTGCAACAAAGAAATGCAAGGCCTGGTTGCCCGAATGAAACAGCATCGTGAGAAGTGCTCCTTCTCAGGAGGAAGCTGCTTTGAAGATGATGAATGGAACATGTCTGAACATGCGGNATNTCTAGGTTAGTAAACTTTCTTATTNCATTCTTTTTACTCAAGGACTGCCTGNCTTCTCTGTTGACATCTTTTGGACGGANTNCTAGATTACTTGAATTCTCATAAAAATGTTTGAGNAAAAGCATANNTGTTATTTTAAGGANCTATCATATTCNNAATGTNATNGTGANNTGAAANGATGACTGAAATAAGCAGACCTTCCTTTTGCAGTTTCACTTCNAAAGTAGTNCTGGNCNCNATGAGCNANNCTAAATGNNCAGTNNNNTAGCGACTNCATTAACTNCTTTTGTTCAGGAGGATCNANCCTGAACATACAGGATTCTGACTGTTCATCTTCAGCATTATCACTGTCCACAGTNTCGAANNTATCTGCTAACGATACNAGTGTTCGAATCNAATGCAAATCGCATATTATACCNCCTGGGGNNNGNTGGGATTGAAATCTCCGTTATCCAGAAACTACCATAGATAAGTTTGCAATAAGAGCCAGCAGATTNCAGCGAGNGGTAATTGATGAAAAAATTGCTCGGTTTGTTTATGCAACAAACTCTCCTTTCCGTATGGTTGAGAACACACGCTTCACTGACGTGGTTCAGTCATTGCGATCAGGATACAGTCCACCCGACAGAGCAGATATAGCAGGCAAATTGCTGGANAAAGTGTAAGAGNAAGAAATTNAACGGGGTNCANAANGCCTAGAGGGAAAAATTGTGAACCTGAGGCNNGATAGGTGGAGCAATGNCCGCAGTGGTCCAATAATATGNGCTTGTGTGACAACAGAANAAGGGNCTNTCTNCCTTNCAGAAACAATCGATACATCAGGAAATGCACGCANAGCAGAATATTTNCAAGAAATAGCAGTAAAAGCTGTAACAAACTGCGAAAAAAAATTCAGATGTCTANTACGCAGCTTCGTCACAGACAACGCTGCAAATGTATCAAAGGTNAGAAGAAATCTAGAANAAAGTGAAGAAAGCCCCAAGCTAATAACATATGGCTGCAGCGCTCATTTGATGCACCTCTTAGCCAAAGACTTCGGTGCTCCAGAAATAAAGGCTAATGTTGCNGAAATTGCAAAATACTTCCATAACAACCANTTTGCAGCAGCTGCTCCGAAGAAAGCAGGAGGAACCAAACTAACTCTCCCACAAGACGTGCGATGGAACTCAGTAGCGGATTGTTTTGNGTGATATATTGAGAACTGGCCTAATCTGACGACAATTTGTGAACAAAATCGTGACAAAATAGATGGAACTGTCACAGCCAAAGTTTNCAANATTGGGCTAAAGAGAAATGTAGAACGTGTGCTGACTATCCCGAAGCCTATTTCCGTAGCCTTGAACAAAGTGCAGGGAAATCGCCGTTTTATTGCTGATGCTGTTGAAATTTGGAAGGNACTGAGTGAGACCTTGAAAAGAGAAATGTNCAATGACAGAGTNAAATTACGGGCATTAAAAAAACGAACGGATCAAGCGCTATNTCCAGCTCATTTTCTTGCCAATATTCTCAATACTCGGTACCAGGGTCAATCCTTAACCGCTGAAGAAGAGTTGGCTATGACATGGGCATCCAGCAATCATCCCTCCATAATACCAACTATAATAAATTTCAGGGCTAAGGGCGAACCATTCAAGAAATATATGTTTGCTGATGAGGCTTTAAAGAAAGTCACGCNAGTGAACTGGTGGAAGTCACTTAAGCACCTGGATCCATCANAGACTACTGAAGTGATAATCCAGCTTTTAACAGCAGTAGCTTCTTCTGCAGGCGTAGAAAGAATATTTTCTTCCTTTGGACTAATTCATTCTAAATTGAGAAATCGCTTGGGAACTGAAAAAGCAGGAAAGCTTGTCTTTCTTTTCCAGTCTATGAATAAACAGGAAGAGGAAGGTGAAGACTGAATTNGCTGCGTAACCCAGTATTTTAAGTTTCTCATGTTGACNTGGCTGAAATAATCTATTTAATTTTTGTNAAAANACACATTTCGCGTTTATAANTAAAATNGTTAAAAATTCAAAAATACACGTGCTTGTTTTGTTAAAATAATTATACACGTTTGTTGTTGAAGAAAAATATCCAAAATACATAAGCTTGTTGTTTTAGTTAAATAAAATAATTTAAATGTCTGTTGCCTTTGGTGACGGTTGTTGTCCTCCNAATACAGNATGGCAAAAATATCCCCCAAATATCCATGATTAATCNANTGAATTGGAGATAGTTCACCTCCCAATGACTTCACAAGTATCTGCTTCAATTAGCTTTGGTAAATAAAATAACCAAATCATAATTCAGTTTTCTGATGTAGCTGCAAAATTAATCTGAAAAAATATCAAAATAAATCACTGCTTTAAAATGTATAGTGTGTACCTTCTAAAAATGAAACCTACATCTATCTCTGAGTTGTGAATAATATGTATTAAGGTTATATCAAACAACAAGAATGCACTTTTTGTAGAAAACAATGATTAAATCGAGGCTTCCTGACCAGTGATTTAAATCGTGATTTAAATCGATTTGATTTAAATCAAATCCACCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| hAT-5_Mam | MCM1 | 812 | 823 | + | 16.12 | CCCGTTTGGGAA |
| hAT-5_Mam | FOXJ2::ELF1 | 2877 | 2887 | + | 16.09 | AAACAGGAAGA |
| hAT-5_Mam | MAZ | 652 | 659 | - | 16.08 | CCCCTCCC |
| hAT-5_Mam | ZNF148 | 650 | 659 | - | 16.08 | CCCCTCCCTC |
| hAT-5_Mam | STAT1::STAT2 | 408 | 420 | + | 16.08 | AGTTTCAGTTCTC |
| hAT-5_Mam | PHL11 | 691 | 700 | - | 16.05 | GGAATATTCT |
| hAT-5_Mam | PHL11 | 691 | 700 | + | 16.03 | AGAATATTCC |
| hAT-5_Mam | PHL7 | 688 | 700 | + | 15.95 | AGCAGAATATTCC |
| hAT-5_Mam | KAN2 | 2776 | 2785 | - | 15.83 | AATATTCTTT |
| hAT-5_Mam | ATHB-15 | 3458 | 3470 | + | 15.82 | AAAACAATGATTA |
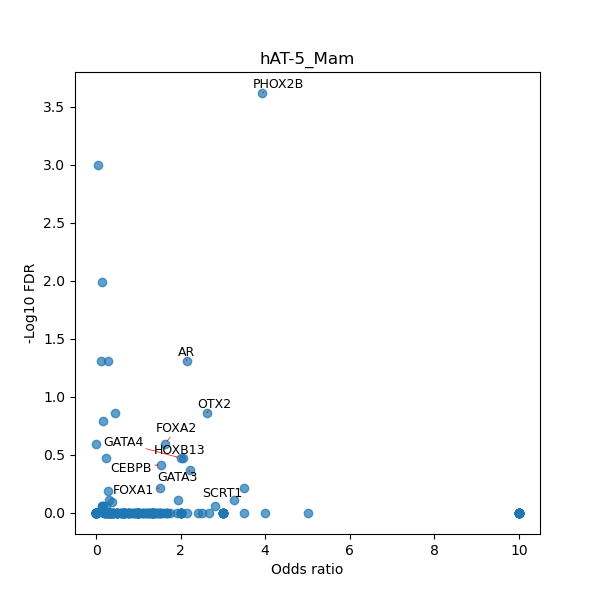
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.