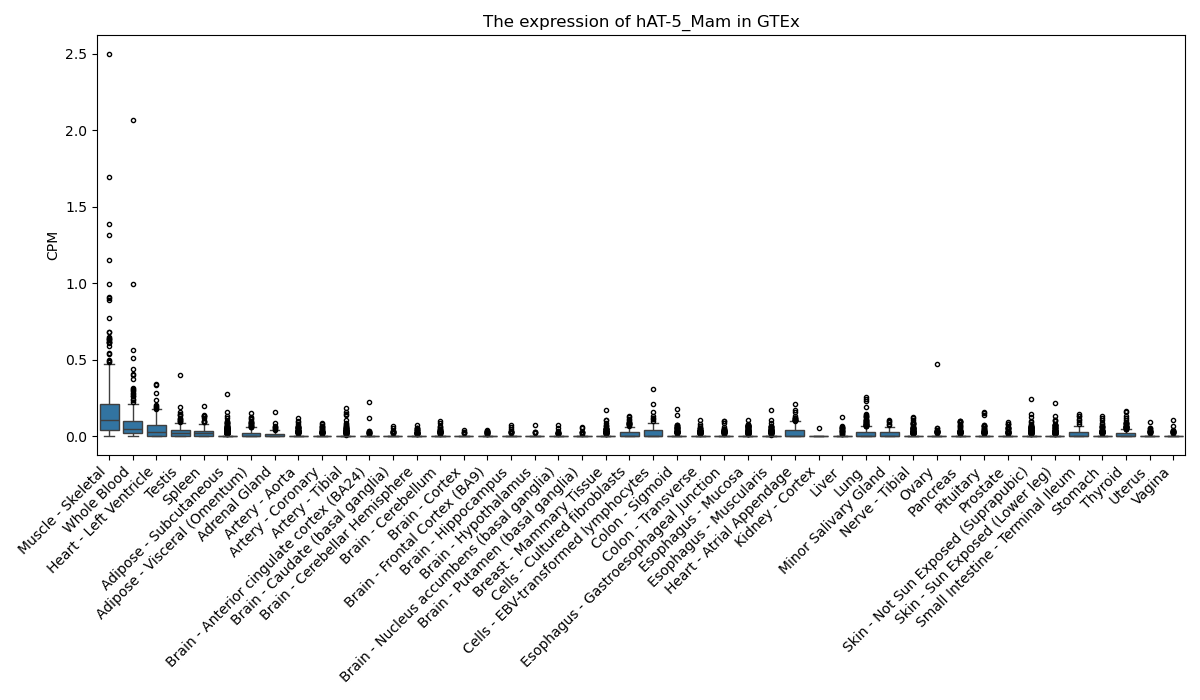
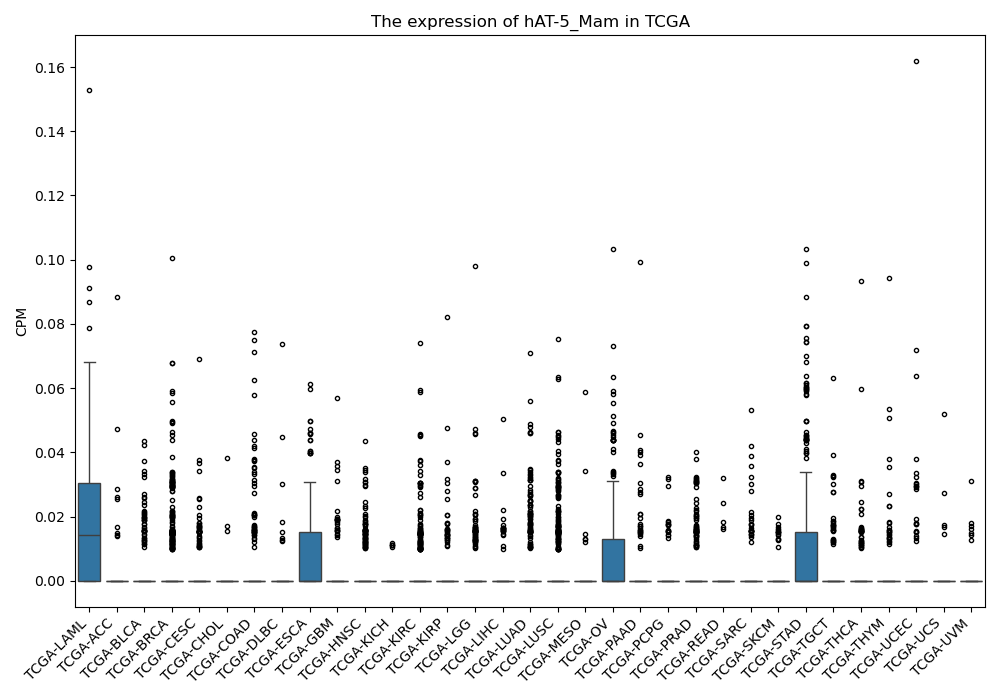
hAT-5_Mam
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001335 |
|---|---|
| TE superfamily | Tag1 |
| TE class | DNA |
| Species | Mammalia |
| Length | 3540 |
| Kimura value | 32.08 |
| Tau index | 0.9753 |
| Description | hAT-Tip100 DNA transposon, hAT-5_Mam subfamily |
| Comment | - |
| Sequence |
CAGGGTAGATAAAAATCNATNATTTTTNAAAAAAAAATTAAAAATCGGATTTTTTAATTTAATCGGATTTTNTANTTTTTNTNGTTAAGATGTTTTNTTAAGAAAAAGAAACCTATATAAAGATAGTTTTAATTTAAGATATGTTAAAGCTCAAAGATATCATCATGGAATAGGGATTATAACTTCTAATTATATANTATTTGAGACAATATATTCATGTAACCTTTTTAGAAAAGTTTTATAAATAGGTCACAACAGGCCACGGACTATATTAGGGGCCCAATCTTATGGGGTTCCCAGAGGCTCCTCTATAGATTANTNAGAAAGATTAAAGAAAACCACTCTACCCAATGGGACTCAGTGCTCAGTCGATCTAGAAGATCCCATTAAGCATCTCTGTGATGCTTAGTTTCAGTTCTCAAACTGTGGATTTGTGTCTCCAGAGATAACATCCTTGTTAACAGCAGTATATGGAGATGAGAGATAACAGACCTGATCAGCCCTATTGTCTCTCTGCAAGTTTGTGTACACAGAGTCAAGCCCTTACCTCTCTCTAAAAGTGCAAAGTTTCAAGAAGTTAAATGAATAGAGGATTGTTGGGGGTGGAGTAGATCTGGGCAAGGAGGAGGAGTCTGGAGATAAATGCAAGGAGGGAGGGGCAGTAGAAACAAAAGAGAAACATTTTCGAGCAGAATATTCCAGAAGTCTTGAGGGATAAGTTTCTGAGTATAGCTTCCATTGATTTGAGATCTAGTGAGGGAATGGTATGGTAGAAGGGAAAACCTATCATGGCAGCAGGCCGTAAAAGAGACCCGTTTGGGAATATTTTAATGAAGTTCCTCTNCCTGTGGGTAAGATAGGCATGCGTGCAAAATGCAAACAGTGCAACAAAGAAATGCAAGGCCTGGTTGCCCGAATGAAACAGCATCGTGAGAAGTGCTCCTTCTCAGGAGGAAGCTGCTTTGAAGATGATGAATGGAACATGTCTGAACATGCGGNATNTCTAGGTTAGTAAACTTTCTTATTNCATTCTTTTTACTCAAGGACTGCCTGNCTTCTCTGTTGACATCTTTTGGACGGANTNCTAGATTACTTGAATTCTCATAAAAATGTTTGAGNAAAAGCATANNTGTTATTTTAAGGANCTATCATATTCNNAATGTNATNGTGANNTGAAANGATGACTGAAATAAGCAGACCTTCCTTTTGCAGTTTCACTTCNAAAGTAGTNCTGGNCNCNATGAGCNANNCTAAATGNNCAGTNNNNTAGCGACTNCATTAACTNCTTTTGTTCAGGAGGATCNANCCTGAACATACAGGATTCTGACTGTTCATCTTCAGCATTATCACTGTCCACAGTNTCGAANNTATCTGCTAACGATACNAGTGTTCGAATCNAATGCAAATCGCATATTATACCNCCTGGGGNNNGNTGGGATTGAAATCTCCGTTATCCAGAAACTACCATAGATAAGTTTGCAATAAGAGCCAGCAGATTNCAGCGAGNGGTAATTGATGAAAAAATTGCTCGGTTTGTTTATGCAACAAACTCTCCTTTCCGTATGGTTGAGAACACACGCTTCACTGACGTGGTTCAGTCATTGCGATCAGGATACAGTCCACCCGACAGAGCAGATATAGCAGGCAAATTGCTGGANAAAGTGTAAGAGNAAGAAATTNAACGGGGTNCANAANGCCTAGAGGGAAAAATTGTGAACCTGAGGCNNGATAGGTGGAGCAATGNCCGCAGTGGTCCAATAATATGNGCTTGTGTGACAACAGAANAAGGGNCTNTCTNCCTTNCAGAAACAATCGATACATCAGGAAATGCACGCANAGCAGAATATTTNCAAGAAATAGCAGTAAAAGCTGTAACAAACTGCGAAAAAAAATTCAGATGTCTANTACGCAGCTTCGTCACAGACAACGCTGCAAATGTATCAAAGGTNAGAAGAAATCTAGAANAAAGTGAAGAAAGCCCCAAGCTAATAACATATGGCTGCAGCGCTCATTTGATGCACCTCTTAGCCAAAGACTTCGGTGCTCCAGAAATAAAGGCTAATGTTGCNGAAATTGCAAAATACTTCCATAACAACCANTTTGCAGCAGCTGCTCCGAAGAAAGCAGGAGGAACCAAACTAACTCTCCCACAAGACGTGCGATGGAACTCAGTAGCGGATTGTTTTGNGTGATATATTGAGAACTGGCCTAATCTGACGACAATTTGTGAACAAAATCGTGACAAAATAGATGGAACTGTCACAGCCAAAGTTTNCAANATTGGGCTAAAGAGAAATGTAGAACGTGTGCTGACTATCCCGAAGCCTATTTCCGTAGCCTTGAACAAAGTGCAGGGAAATCGCCGTTTTATTGCTGATGCTGTTGAAATTTGGAAGGNACTGAGTGAGACCTTGAAAAGAGAAATGTNCAATGACAGAGTNAAATTACGGGCATTAAAAAAACGAACGGATCAAGCGCTATNTCCAGCTCATTTTCTTGCCAATATTCTCAATACTCGGTACCAGGGTCAATCCTTAACCGCTGAAGAAGAGTTGGCTATGACATGGGCATCCAGCAATCATCCCTCCATAATACCAACTATAATAAATTTCAGGGCTAAGGGCGAACCATTCAAGAAATATATGTTTGCTGATGAGGCTTTAAAGAAAGTCACGCNAGTGAACTGGTGGAAGTCACTTAAGCACCTGGATCCATCANAGACTACTGAAGTGATAATCCAGCTTTTAACAGCAGTAGCTTCTTCTGCAGGCGTAGAAAGAATATTTTCTTCCTTTGGACTAATTCATTCTAAATTGAGAAATCGCTTGGGAACTGAAAAAGCAGGAAAGCTTGTCTTTCTTTTCCAGTCTATGAATAAACAGGAAGAGGAAGGTGAAGACTGAATTNGCTGCGTAACCCAGTATTTTAAGTTTCTCATGTTGACNTGGCTGAAATAATCTATTTAATTTTTGTNAAAANACACATTTCGCGTTTATAANTAAAATNGTTAAAAATTCAAAAATACACGTGCTTGTTTTGTTAAAATAATTATACACGTTTGTTGTTGAAGAAAAATATCCAAAATACATAAGCTTGTTGTTTTAGTTAAATAAAATAATTTAAATGTCTGTTGCCTTTGGTGACGGTTGTTGTCCTCCNAATACAGNATGGCAAAAATATCCCCCAAATATCCATGATTAATCNANTGAATTGGAGATAGTTCACCTCCCAATGACTTCACAAGTATCTGCTTCAATTAGCTTTGGTAAATAAAATAACCAAATCATAATTCAGTTTTCTGATGTAGCTGCAAAATTAATCTGAAAAAATATCAAAATAAATCACTGCTTTAAAATGTATAGTGTGTACCTTCTAAAAATGAAACCTACATCTATCTCTGAGTTGTGAATAATATGTATTAAGGTTATATCAAACAACAAGAATGCACTTTTTGTAGAAAACAATGATTAAATCGAGGCTTCCTGACCAGTGATTTAAATCGTGATTTAAATCGATTTGATTTAAATCAAATCCACCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| hAT-5_Mam | FOXO1::ELK1 | 2875 | 2887 | + | 17.34 | ATAAACAGGAAGA |
| hAT-5_Mam | TP53 | 980 | 997 | + | 17.29 | AACATGTCTGAACATGCG |
| hAT-5_Mam | FOXO1::ELF1 | 2875 | 2887 | + | 17.28 | ATAAACAGGAAGA |
| hAT-5_Mam | HSF4 | 692 | 704 | - | 17.10 | TTCTGGAATATTC |
| hAT-5_Mam | SOL1 | 3007 | 3021 | + | 17.00 | GTTAAAAATTCAAAA |
| hAT-5_Mam | Ascl2 | 2105 | 2114 | + | 16.91 | AGCAGCTGCT |
| hAT-5_Mam | Ascl2 | 2105 | 2114 | - | 16.91 | AGCAGCTGCT |
| hAT-5_Mam | Spi1 | 2877 | 2889 | + | 16.91 | AAACAGGAAGAGG |
| hAT-5_Mam | FOXO1::ELK3 | 2875 | 2887 | + | 16.91 | ATAAACAGGAAGA |
| hAT-5_Mam | sc | 2105 | 2114 | + | 16.80 | AGCAGCTGCT |
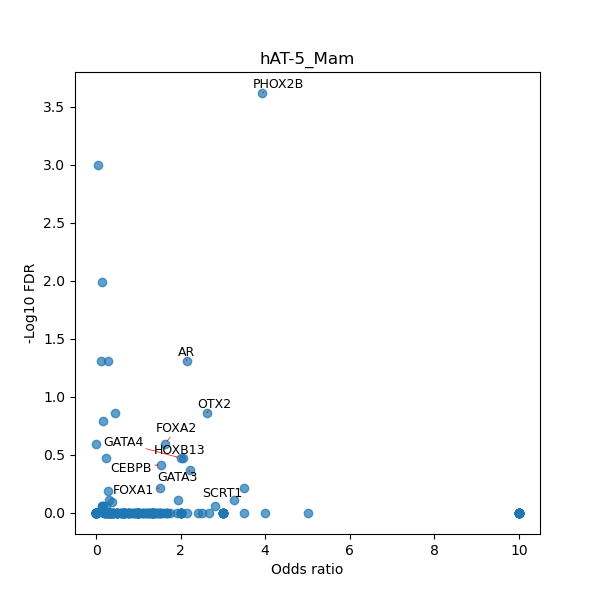
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.