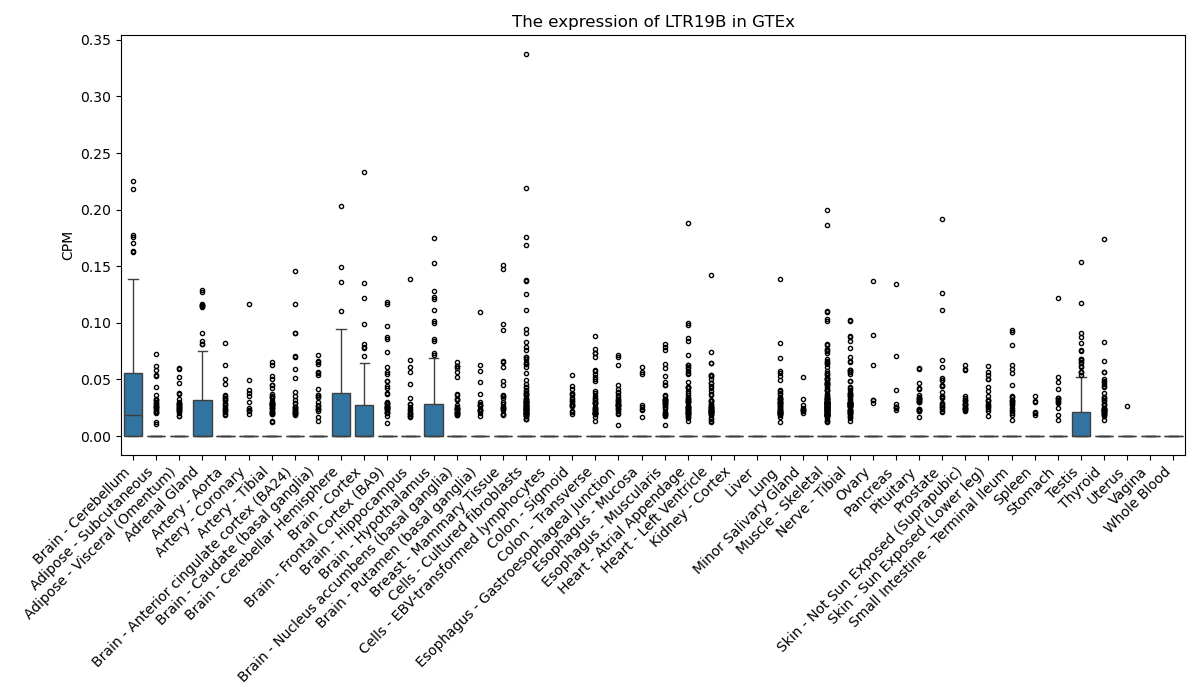
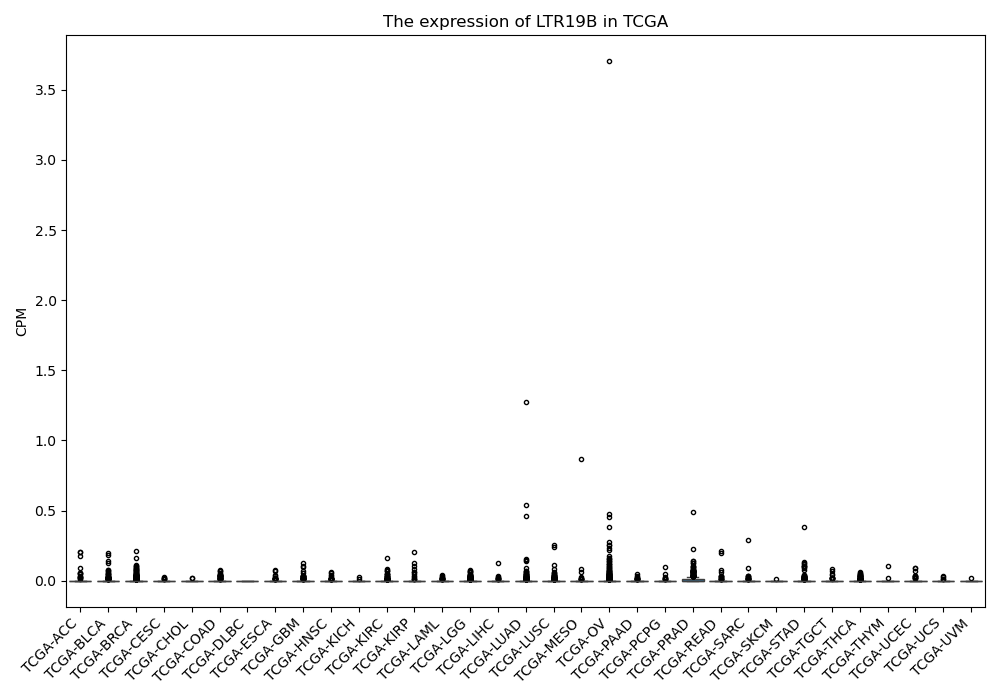
LTR19B
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000432 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Haplorrhini |
| Length | 580 |
| Kimura value | 7.55 |
| Tau index | 1.0000 |
| Description | LTR19B (Long Terminal Repeat) for ERV1 endogenous retrovirus |
| Comment | LTR19 elements are flanked by 5 bp target site duplication. |
| Sequence |
TGACAGAGCAGGAGCACCGTCATCTCGGACAAACACCGCCACTTTAAGTTCCAGCTCCCTTTCTAGCCTCATGCATTTCAAGGAAATCACTTCTCTTCTAACTACAAGCAGCCAGAAAGAGCAGACAGTAAAACACAGATAAGACAGCTCGGGCACAGAGGGAGGTGGGGGGAAAGTCTCTTGGGTAACTGCCAAACTTCACCCTCATACAATGGGCCCCAGTAAAACAGTGGGCCTTAATAAGCACATTCCTTTCCCTTCAGGTGCACTAAGATAGGGAAGCTAAAAGCAGACTCGGGGNNNCGNCGGGGGGTATGCCTGCAGCTGCAGAAAGATGTATGGGAACAGACACACAACTCTCCCTCCCAGATAAGCACAACAAAGAGACACAGAAGCAGTCCAAGCCTCTGATAAACTCTCCCACCCTGAATCCTTAAAAACTCTTAGTCTGTAAGAGAGTGTGCCTCTGACCTAACTCGGCCAGAAGCCCCTCTCAGGTTTGTTTTCTCTAAAATAAACCTGTCTTTGNCGACTGTCGAGCCACCTTTCGTGTTTCTTTCCTCTTTCTTTAATTCTTACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR19B | BPC1 | 154 | 177 | + | 1.18 | CACAGAGGGAGGTGGGGGGAAAGT |
| LTR19B | TRP2 | 423 | 443 | + | -0.59 | ACCCTGAATCCTTAAAAACTC |
| LTR19B | BPC1 | 152 | 175 | + | -1.35 | GGCACAGAGGGAGGTGGGGGGAAA |
| LTR19B | BPC1 | 556 | 579 | - | -1.68 | GTAAGAATTAAAGAAAGAGGAAAG |
| LTR19B | BPC6 | 555 | 575 | + | -1.97 | TCTTTCCTCTTTCTTTAATTC |
| LTR19B | E2F7 | 163 | 176 | - | -5.61 | CTTTCCCCCCACCT |
| LTR19B | EWSR1-FLI1 | 544 | 561 | - | -13.14 | GGAAAGAAACACGAAAGG |
| LTR19B | EWSR1-FLI1 | 152 | 169 | + | -16.00 | GGCACAGAGGGAGGTGGG |
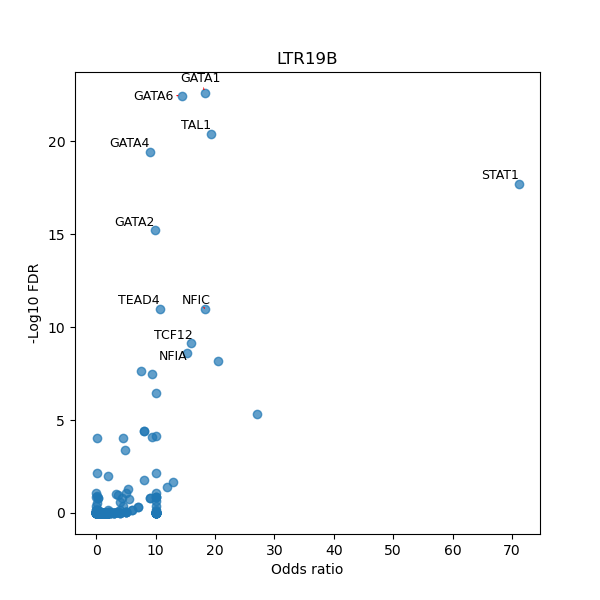
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.