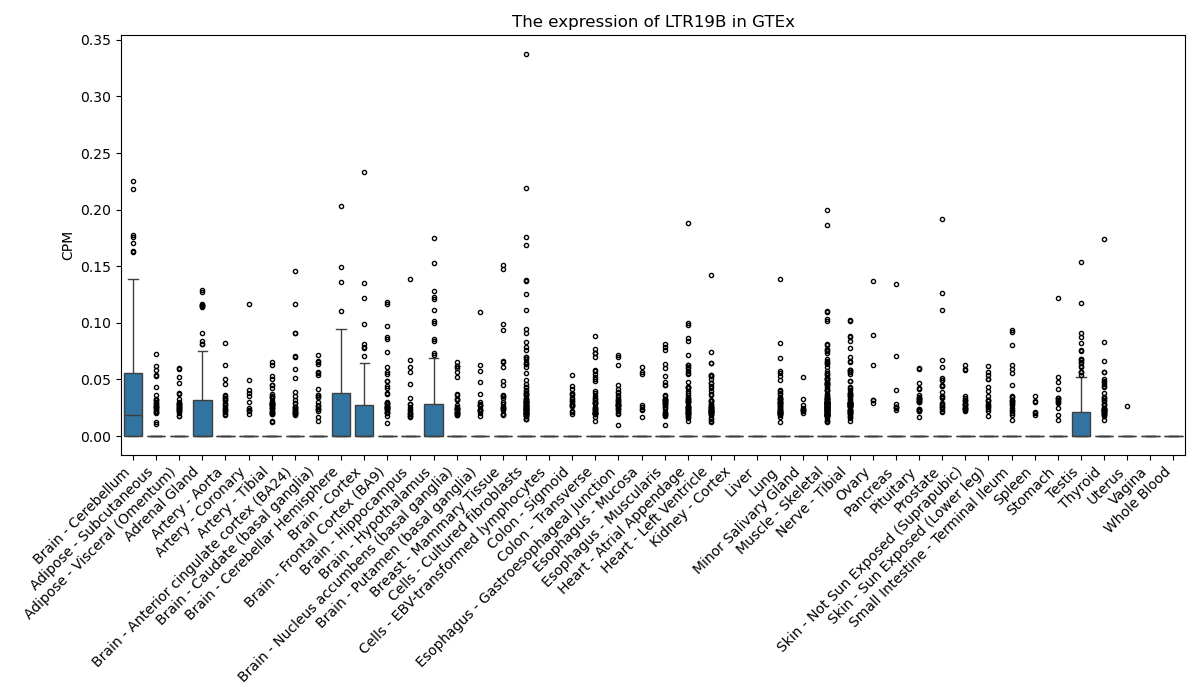
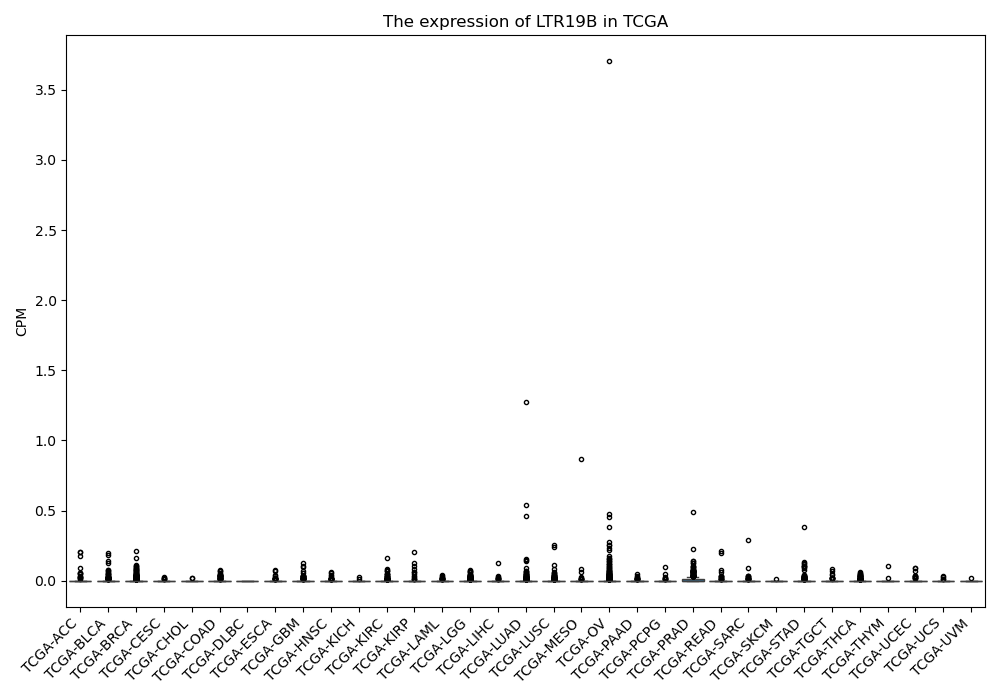
LTR19B
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000432 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Haplorrhini |
| Length | 580 |
| Kimura value | 7.55 |
| Tau index | 1.0000 |
| Description | LTR19B (Long Terminal Repeat) for ERV1 endogenous retrovirus |
| Comment | LTR19 elements are flanked by 5 bp target site duplication. |
| Sequence |
TGACAGAGCAGGAGCACCGTCATCTCGGACAAACACCGCCACTTTAAGTTCCAGCTCCCTTTCTAGCCTCATGCATTTCAAGGAAATCACTTCTCTTCTAACTACAAGCAGCCAGAAAGAGCAGACAGTAAAACACAGATAAGACAGCTCGGGCACAGAGGGAGGTGGGGGGAAAGTCTCTTGGGTAACTGCCAAACTTCACCCTCATACAATGGGCCCCAGTAAAACAGTGGGCCTTAATAAGCACATTCCTTTCCCTTCAGGTGCACTAAGATAGGGAAGCTAAAAGCAGACTCGGGGNNNCGNCGGGGGGTATGCCTGCAGCTGCAGAAAGATGTATGGGAACAGACACACAACTCTCCCTCCCAGATAAGCACAACAAAGAGACACAGAAGCAGTCCAAGCCTCTGATAAACTCTCCCACCCTGAATCCTTAAAAACTCTTAGTCTGTAAGAGAGTGTGCCTCTGACCTAACTCGGCCAGAAGCCCCTCTCAGGTTTGTTTTCTCTAAAATAAACCTGTCTTTGNCGACTGTCGAGCCACCTTTCGTGTTTCTTTCCTCTTTCTTTAATTCTTACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR19B | ZNF418 | 277 | 291 | + | 21.12 | GGGAAGCTAAAAGCA |
| LTR19B | eor-1 | 383 | 395 | + | 15.53 | AGAGACACAGAAG |
| LTR19B | PATZ1 | 161 | 171 | + | 15.46 | GGAGGTGGGGG |
| LTR19B | ac | 321 | 328 | + | 14.47 | GCAGCTGC |
| LTR19B | ac | 321 | 328 | - | 14.47 | GCAGCTGC |
| LTR19B | ZNF766 | 513 | 521 | + | 14.38 | AATAAACCT |
| LTR19B | MYOG | 321 | 328 | + | 14.35 | GCAGCTGC |
| LTR19B | MYOG | 321 | 328 | - | 14.35 | GCAGCTGC |
| LTR19B | AFT1 | 262 | 271 | - | 14.32 | TAGTGCACCT |
| LTR19B | Mecom | 137 | 147 | + | 14.20 | AGATAAGACAG |
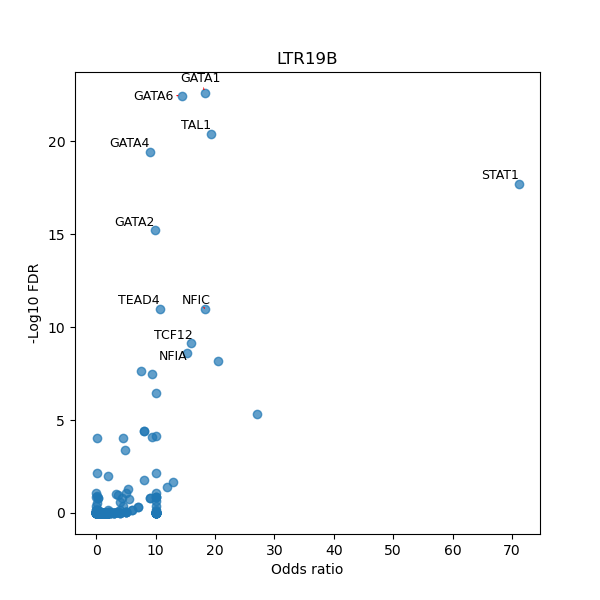
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.