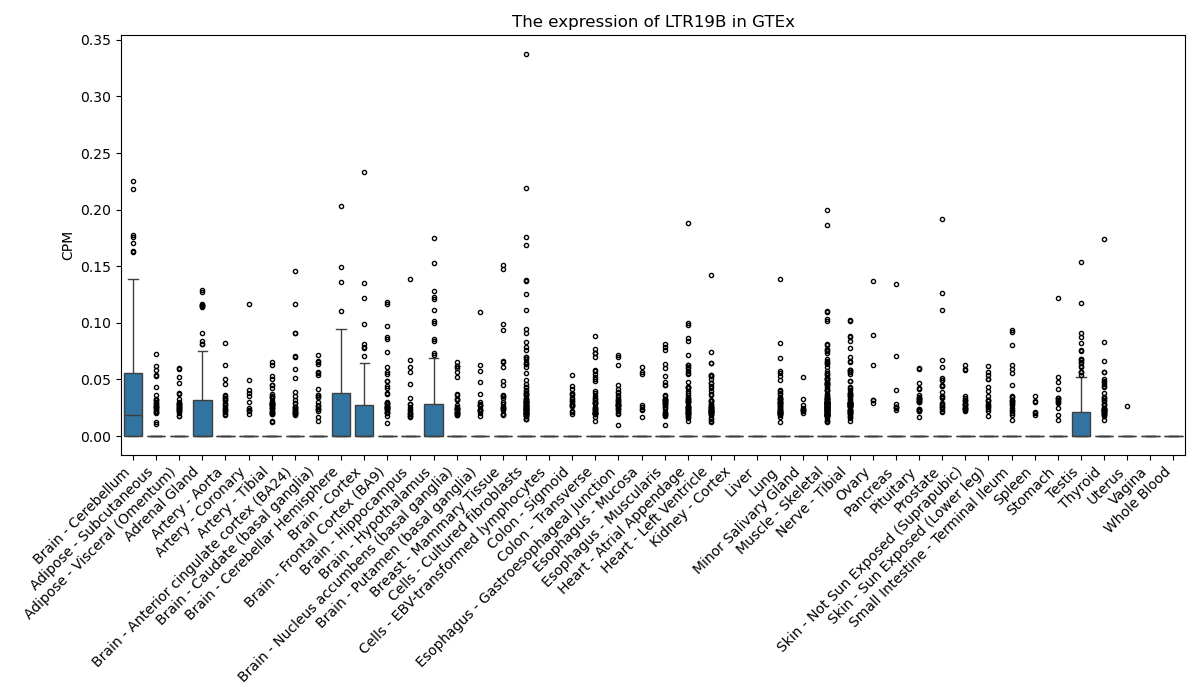
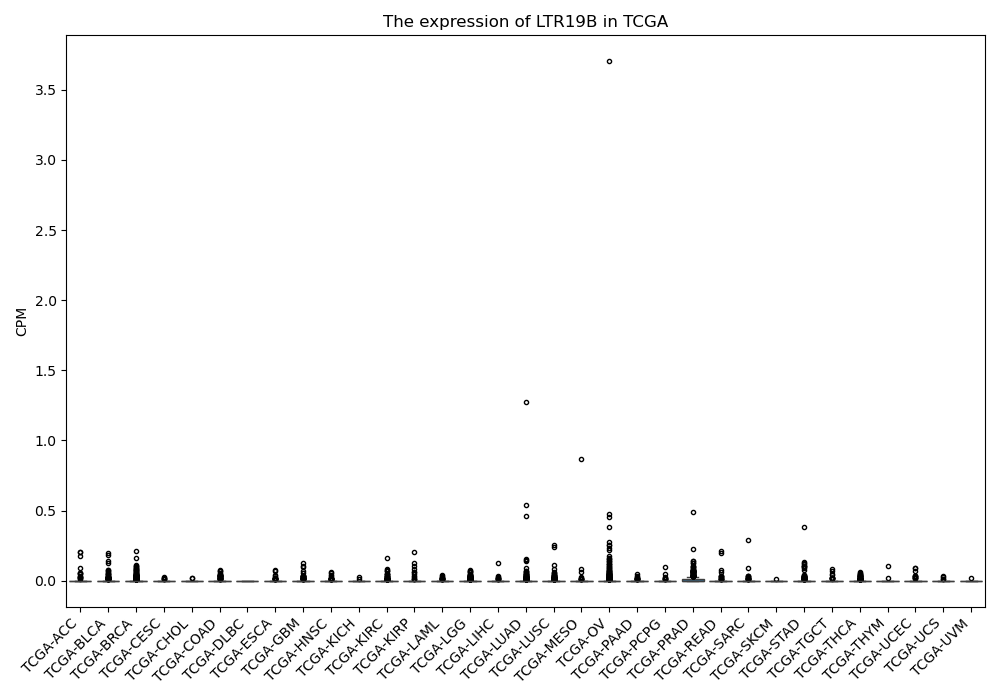
LTR19B
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000432 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Haplorrhini |
| Length | 580 |
| Kimura value | 7.55 |
| Tau index | 1.0000 |
| Description | LTR19B (Long Terminal Repeat) for ERV1 endogenous retrovirus |
| Comment | LTR19 elements are flanked by 5 bp target site duplication. |
| Sequence |
TGACAGAGCAGGAGCACCGTCATCTCGGACAAACACCGCCACTTTAAGTTCCAGCTCCCTTTCTAGCCTCATGCATTTCAAGGAAATCACTTCTCTTCTAACTACAAGCAGCCAGAAAGAGCAGACAGTAAAACACAGATAAGACAGCTCGGGCACAGAGGGAGGTGGGGGGAAAGTCTCTTGGGTAACTGCCAAACTTCACCCTCATACAATGGGCCCCAGTAAAACAGTGGGCCTTAATAAGCACATTCCTTTCCCTTCAGGTGCACTAAGATAGGGAAGCTAAAAGCAGACTCGGGGNNNCGNCGGGGGGTATGCCTGCAGCTGCAGAAAGATGTATGGGAACAGACACACAACTCTCCCTCCCAGATAAGCACAACAAAGAGACACAGAAGCAGTCCAAGCCTCTGATAAACTCTCCCACCCTGAATCCTTAAAAACTCTTAGTCTGTAAGAGAGTGTGCCTCTGACCTAACTCGGCCAGAAGCCCCTCTCAGGTTTGTTTTCTCTAAAATAAACCTGTCTTTGNCGACTGTCGAGCCACCTTTCGTGTTTCTTTCCTCTTTCTTTAATTCTTACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR19B | OSR2 | 389 | 396 | + | 14.17 | ACAGAAGC |
| LTR19B | MYB49 | 468 | 478 | + | 13.99 | TGACCTAACTC |
| LTR19B | DREB2A | 34 | 42 | - | 13.95 | GTGGCGGTG |
| LTR19B | ZFP14 | 247 | 261 | - | 13.91 | GAAGGGAAAGGAATG |
| LTR19B | PRDM9 | 157 | 176 | + | 13.87 | AGAGGGAGGTGGGGGGAAAG |
| LTR19B | GATA6 | 137 | 144 | - | 13.85 | TCTTATCT |
| LTR19B | elt-1 | 137 | 144 | - | 13.78 | TCTTATCT |
| LTR19B | PI | 556 | 569 | - | 13.76 | AAGAAAGAGGAAAG |
| LTR19B | Gata3 | 137 | 144 | - | 13.74 | TCTTATCT |
| LTR19B | TRPS1 | 137 | 144 | - | 13.71 | TCTTATCT |
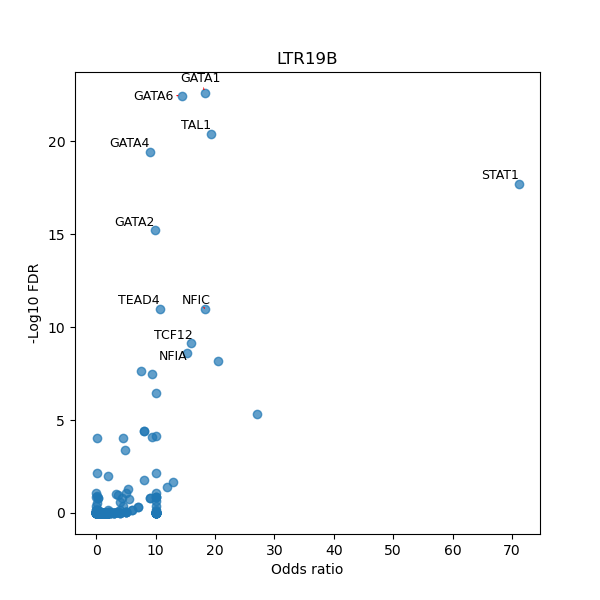
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.