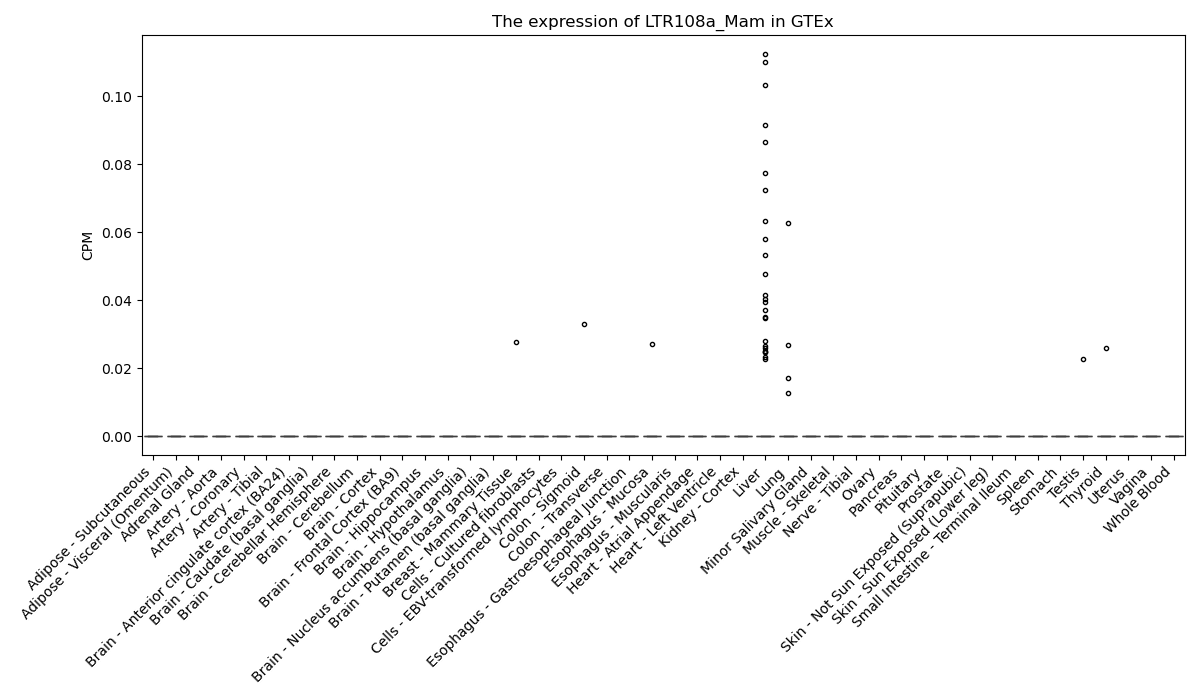
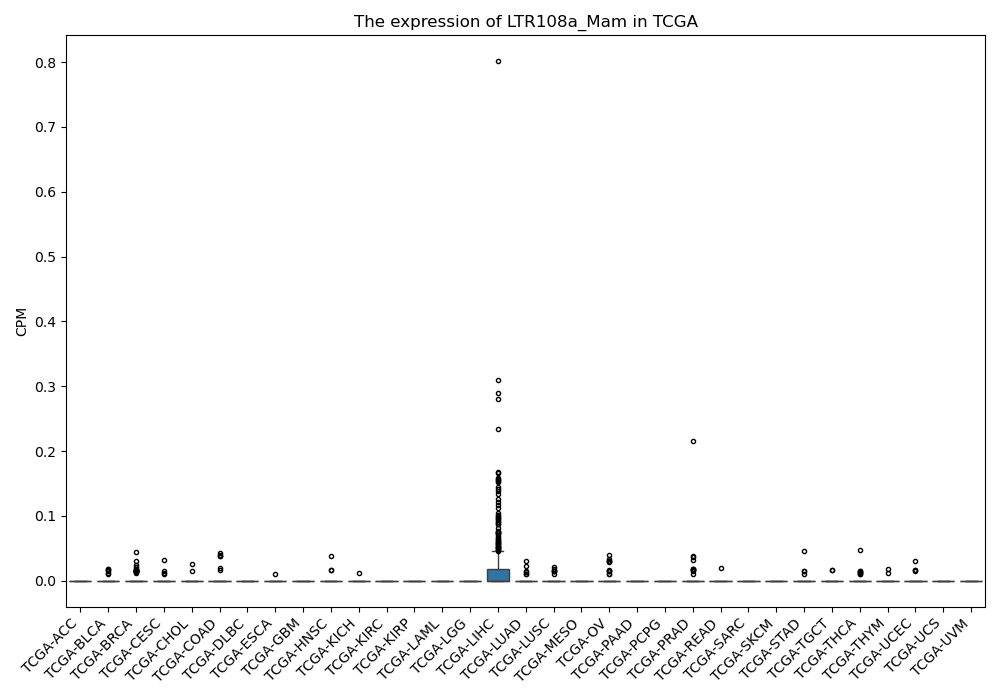
LTR108a_Mam
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000384 |
|---|---|
| TE superfamily | ERV3 |
| TE class | LTR |
| Species | Eutheria |
| Length | 511 |
| Kimura value | 24.95 |
| Tau index | 0.0000 |
| Description | LTR108a_Mam (Long Terminal Repeat) for ERVL endogenous retrovirus |
| Comment | LTR108a_Mam has 5 bp TSDs, with a preference for GNNNC. The substitution level found in the Laurasiatherian reconstruction is ~21%. |
| Sequence |
TGTGTAGGATAGAGTTGATTTACAGCTGGACTGGGAAAGCTCTATCTTTATCTCTGGGGACGCCTGGATGTCTGGCTTTGACTCTTGGCCAAGATAAGGGCAGTGGGATTTATGATGTGCTGTTTAAAGTCCGAAACTCCCCTGGCGACCCCCAGGGTCAGTTTGTTTACTGATGGTTTTCANCTGGCNTAACCGGTTAGACAGCCTGGCTTGACTACGTGACCAGAGGGGCATAAAAGACCCCGCTGGGAACTTCTGCCTTGAGCTCTCCCGAAGCCAAGATTCCTCGCACAGATCTTGGTGGTTCAATCCGGAGACGAAGCGCGTCCTGTGTGCGAATAAGGACCCTGGGGAGCTTGCGCCTGGATGTCTCTCGGACCCCCAATGCTTGCCTGCACTCTCTTTCTCTTGCTGCACCGTATCCTTTGCCTTTAAATAAAAGCCTTACGTGAGTATGCTCTGTGGAGTCTGGTGAGTCCTTTCAAATATCCGAACTTGNGAAATCTTTGCA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR108a_Mam | br | 162 | 172 | - | 13.41 | CAGTAAACAAA |
| LTR108a_Mam | FoxP | 163 | 171 | - | 13.39 | AGTAAACAA |
| LTR108a_Mam | D1 | 432 | 439 | + | 13.38 | TTAAATAA |
| LTR108a_Mam | VIP1 | 20 | 29 | + | 13.38 | TTACAGCTGG |
| LTR108a_Mam | GRHL1 | 190 | 199 | + | 13.35 | TAACCGGTTA |
| LTR108a_Mam | GRHL1 | 190 | 199 | - | 13.35 | TAACCGGTTA |
| LTR108a_Mam | Foxj2 | 163 | 170 | - | 13.32 | GTAAACAA |
| LTR108a_Mam | PRDM1 | 402 | 408 | + | 13.28 | CTTTCTC |
| LTR108a_Mam | Ebf4 | 138 | 148 | - | 13.25 | TCGCCAGGGGA |
| LTR108a_Mam | HRS1 | 275 | 286 | + | 13.24 | AGCCAAGATTCC |
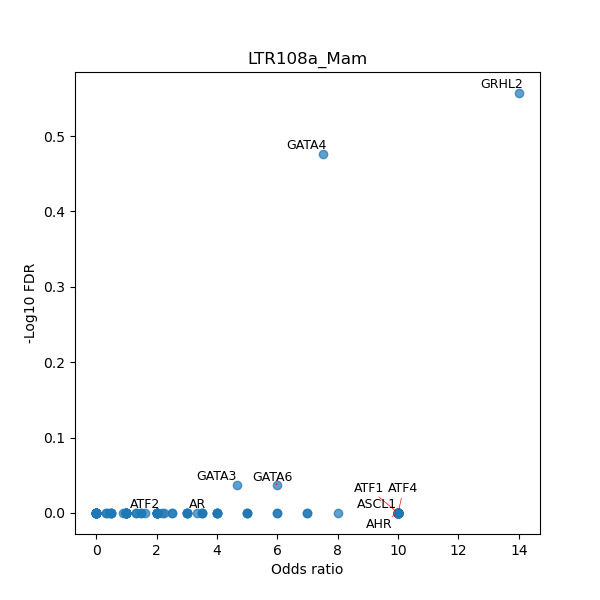
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.