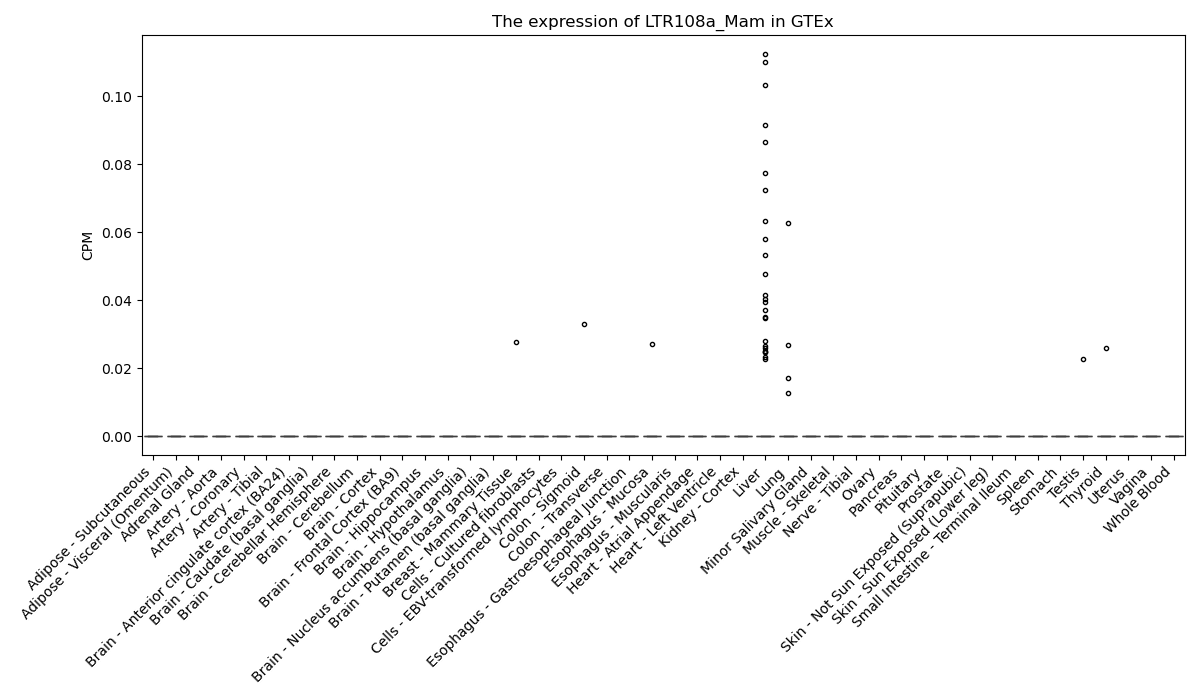
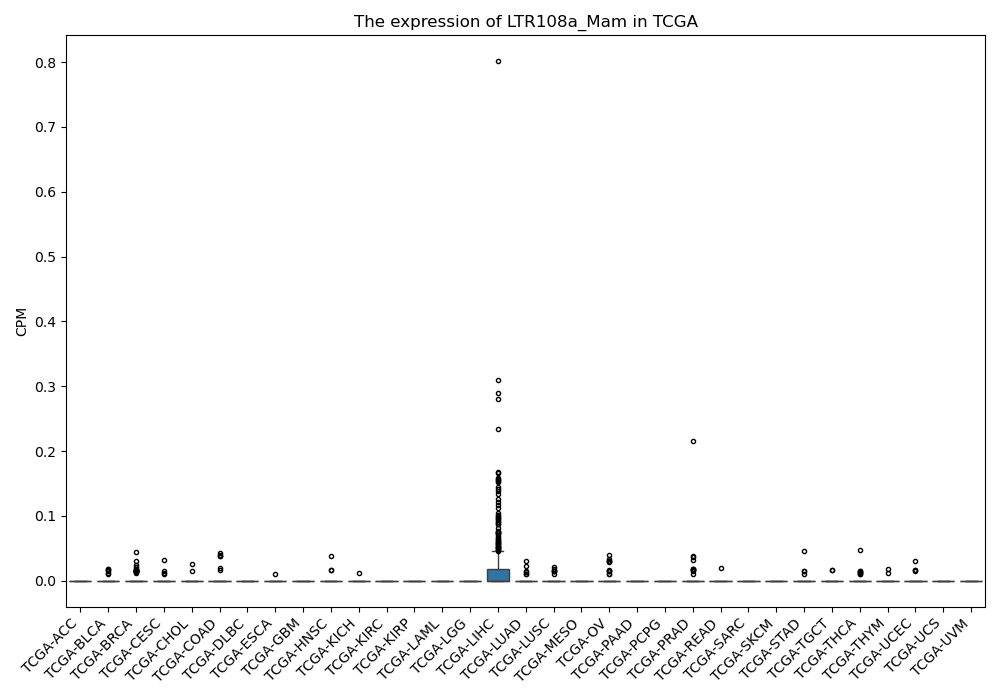
LTR108a_Mam
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000384 |
|---|---|
| TE superfamily | ERV3 |
| TE class | LTR |
| Species | Eutheria |
| Length | 511 |
| Kimura value | 24.95 |
| Tau index | 0.0000 |
| Description | LTR108a_Mam (Long Terminal Repeat) for ERVL endogenous retrovirus |
| Comment | LTR108a_Mam has 5 bp TSDs, with a preference for GNNNC. The substitution level found in the Laurasiatherian reconstruction is ~21%. |
| Sequence |
TGTGTAGGATAGAGTTGATTTACAGCTGGACTGGGAAAGCTCTATCTTTATCTCTGGGGACGCCTGGATGTCTGGCTTTGACTCTTGGCCAAGATAAGGGCAGTGGGATTTATGATGTGCTGTTTAAAGTCCGAAACTCCCCTGGCGACCCCCAGGGTCAGTTTGTTTACTGATGGTTTTCANCTGGCNTAACCGGTTAGACAGCCTGGCTTGACTACGTGACCAGAGGGGCATAAAAGACCCCGCTGGGAACTTCTGCCTTGAGCTCTCCCGAAGCCAAGATTCCTCGCACAGATCTTGGTGGTTCAATCCGGAGACGAAGCGCGTCCTGTGTGCGAATAAGGACCCTGGGGAGCTTGCGCCTGGATGTCTCTCGGACCCCCAATGCTTGCCTGCACTCTCTTTCTCTTGCTGCACCGTATCCTTTGCCTTTAAATAAAAGCCTTACGTGAGTATGCTCTGTGGAGTCTGGTGAGTCCTTTCAAATATCCGAACTTGNGAAATCTTTGCA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR108a_Mam | ZNF524 | 373 | 381 | + | 15.60 | CTCGGACCC |
| LTR108a_Mam | SUT2 | 126 | 135 | + | 15.59 | AAAGTCCGAA |
| LTR108a_Mam | FOXN3 | 163 | 170 | - | 15.19 | GTAAACAA |
| LTR108a_Mam | Foxj3 | 162 | 170 | - | 15.06 | GTAAACAAA |
| LTR108a_Mam | CHES-1-like | 162 | 171 | - | 14.87 | AGTAAACAAA |
| LTR108a_Mam | kn | 51 | 60 | - | 14.39 | TCCCCAGAGA |
| LTR108a_Mam | FOXP2 | 163 | 171 | - | 14.35 | AGTAAACAA |
| LTR108a_Mam | NRG1 | 343 | 350 | - | 14.28 | CAGGGTCC |
| LTR108a_Mam | kn | 345 | 354 | - | 14.26 | TCCCCAGGGT |
| LTR108a_Mam | GATA4 | 92 | 99 | - | 14.20 | CCTTATCT |
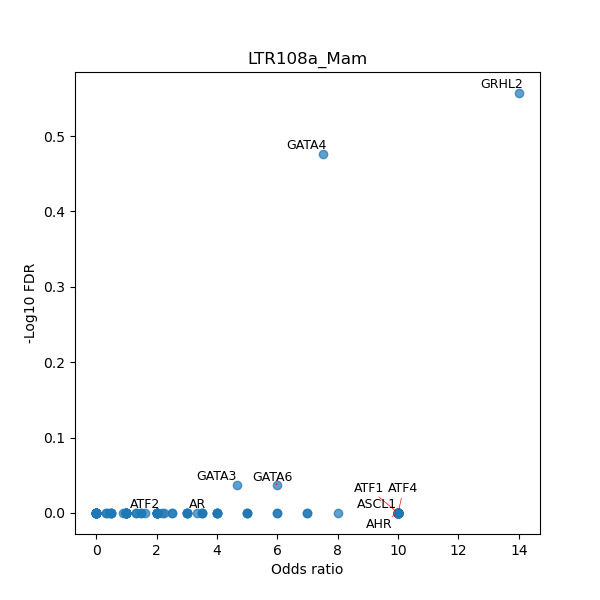
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.