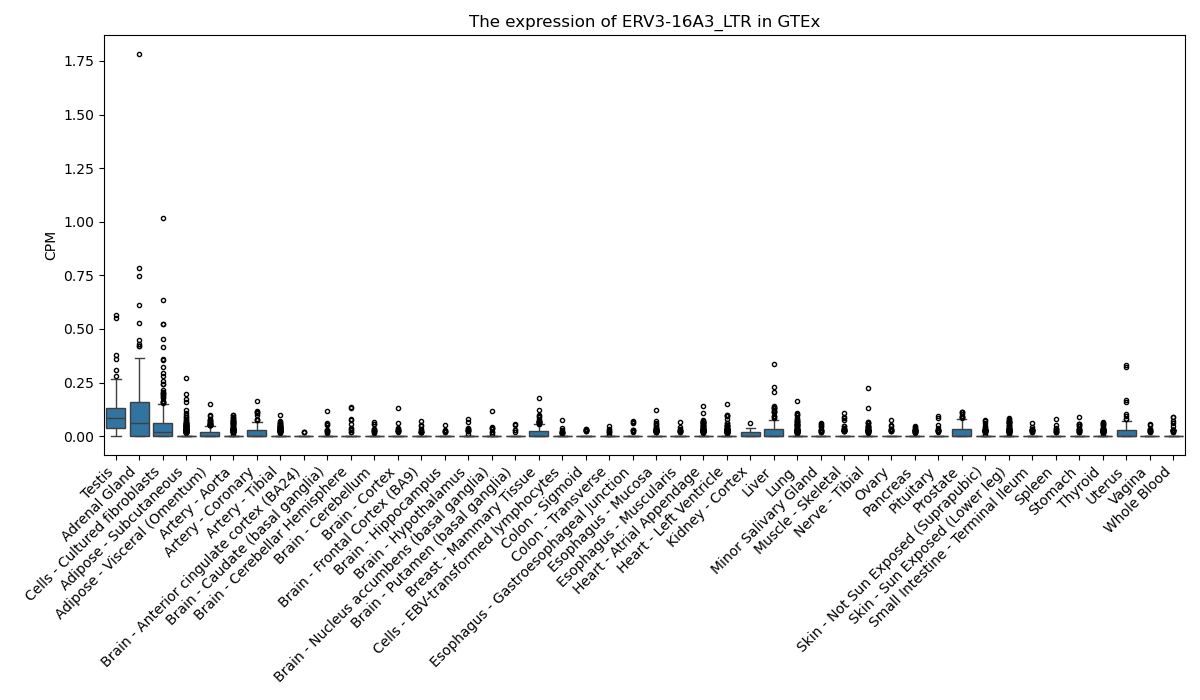
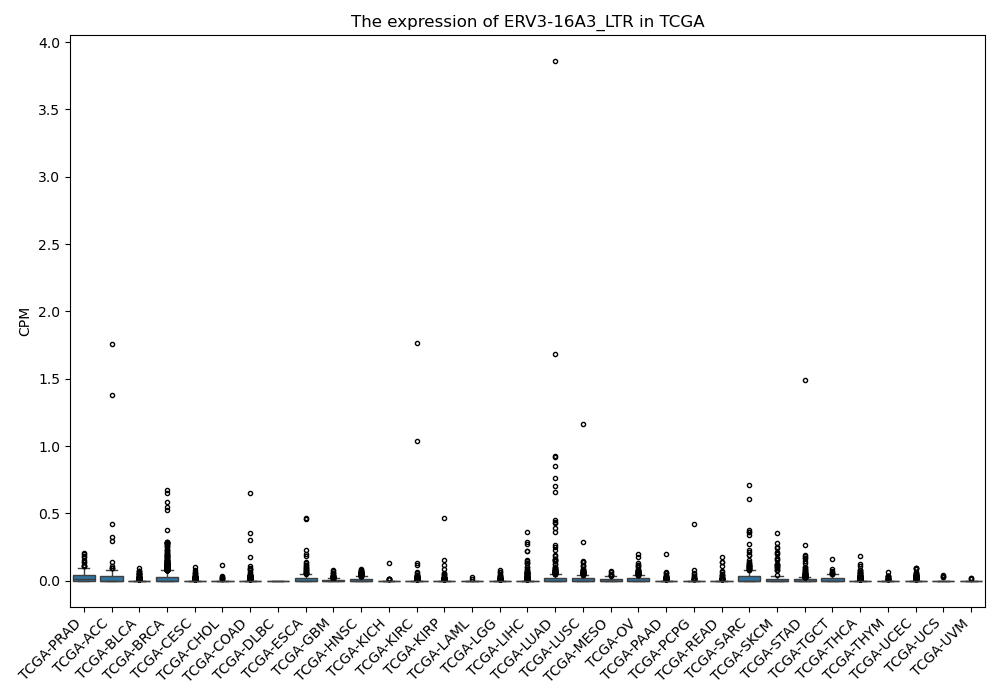
ERV3-16A3_LTR
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000117 |
|---|---|
| TE superfamily | ERV3 |
| TE class | LTR |
| Species | Eutheria |
| Length | 422 |
| Kimura value | 26.96 |
| Tau index | 0.9781 |
| Description | Long terminal repeat, ERV3-16A3_LTR subfamily |
| Comment | Associated with ERV3-16A3_I. |
| Sequence |
TGTGGCAGCCACGGAGGTGCGCCGCTCGGATCTCCCTTCAAGAAAGAACTTGCCGTTCAGCTGCGAGGAGTGCGGTTAGCTGACAGCCTCCAGCTGTTAGCGCCTTCAGGATCCGCCTCAGCTTTCGAGCCGAGGCCACGCTCTTCCCGGGCAGCCCCCAGCCAATGACTGAGCANGGCGGGGGTACNAGGGCCTGGCCATTTCTGCCCAACGCGGGACTCCTCTAACGGGCAATCTTTGCTCCGGAAGAGCTCCCCGTTGGGTTGGCCGAGACTTTGTCAGATCTGCATCGCGGTCTGAGGCTCTCCCTGCCCAATCCTGCTTCCTCTCCTTTTNTCTTTCACAGGCGTTACCCCCCAATAAACCTCTTGCACTCCTAACTCCGTCTCAGCGTCTGCTTCCCGGAGGACCCAACTGACACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| ERV3-16A3_LTR | URC2 | 378 | 385 | - | 12.98 | CGGAGTTA |
| ERV3-16A3_LTR | NAC078 | 39 | 45 | + | 12.96 | CAAGAAA |
| ERV3-16A3_LTR | GLYMA-07G038400 | 415 | 421 | + | 12.89 | CTGACAC |
| ERV3-16A3_LTR | ETV5::FOXO1 | 317 | 326 | - | 12.89 | GGAAGCAGGA |
| ERV3-16A3_LTR | BZIP52 | 81 | 88 | + | 12.86 | TGACAGCC |
| ERV3-16A3_LTR | Atoh7 | 91 | 97 | + | 12.78 | CAGCTGT |
| ERV3-16A3_LTR | Bhlha15 | 90 | 97 | - | 12.75 | ACAGCTGG |
| ERV3-16A3_LTR | Bhlha15 | 90 | 97 | + | 12.75 | CCAGCTGT |
| ERV3-16A3_LTR | ELK4 | 243 | 251 | - | 12.75 | CTCTTCCGG |
| ERV3-16A3_LTR | Hoxd13 | 358 | 364 | + | 12.73 | CAATAAA |
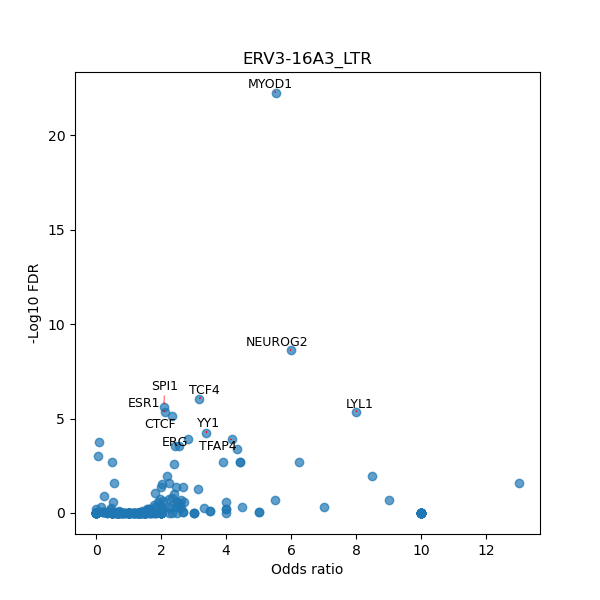
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.