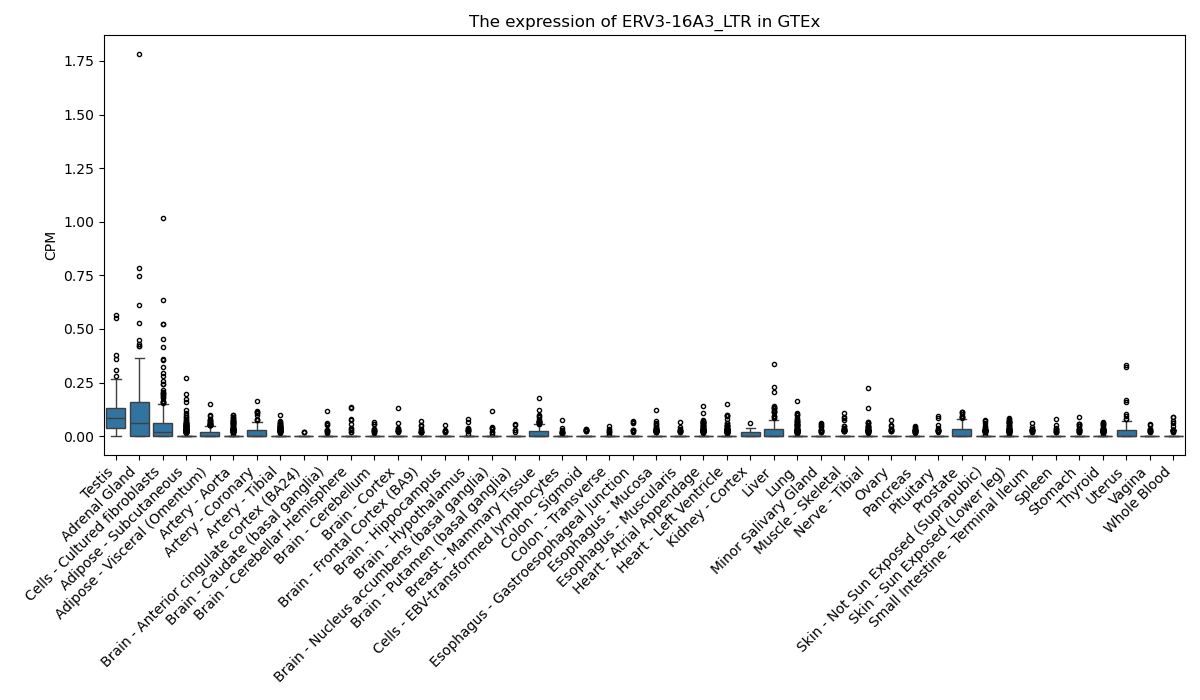
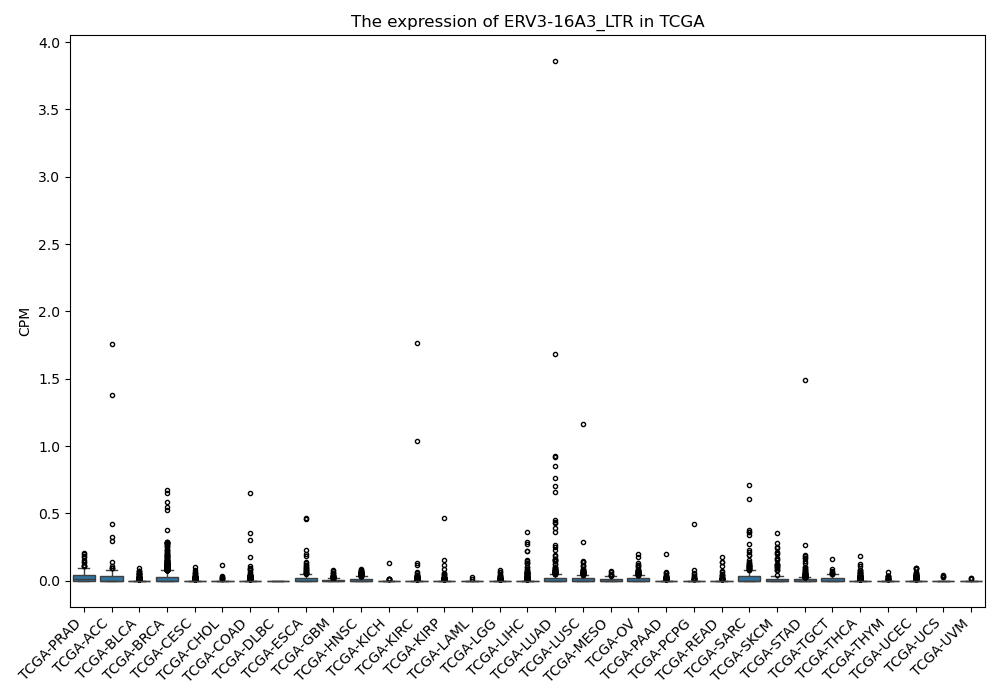
ERV3-16A3_LTR
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000117 |
|---|---|
| TE superfamily | ERV3 |
| TE class | LTR |
| Species | Eutheria |
| Length | 422 |
| Kimura value | 26.96 |
| Tau index | 0.9781 |
| Description | Long terminal repeat, ERV3-16A3_LTR subfamily |
| Comment | Associated with ERV3-16A3_I. |
| Sequence |
TGTGGCAGCCACGGAGGTGCGCCGCTCGGATCTCCCTTCAAGAAAGAACTTGCCGTTCAGCTGCGAGGAGTGCGGTTAGCTGACAGCCTCCAGCTGTTAGCGCCTTCAGGATCCGCCTCAGCTTTCGAGCCGAGGCCACGCTCTTCCCGGGCAGCCCCCAGCCAATGACTGAGCANGGCGGGGGTACNAGGGCCTGGCCATTTCTGCCCAACGCGGGACTCCTCTAACGGGCAATCTTTGCTCCGGAAGAGCTCCCCGTTGGGTTGGCCGAGACTTTGTCAGATCTGCATCGCGGTCTGAGGCTCTCCCTGCCCAATCCTGCTTCCTCTCCTTTTNTCTTTCACAGGCGTTACCCCCCAATAAACCTCTTGCACTCCTAACTCCGTCTCAGCGTCTGCTTCCCGGAGGACCCAACTGACACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| ERV3-16A3_LTR | Nfe2l2 | 165 | 175 | + | 15.97 | ATGACTGAGCA |
| ERV3-16A3_LTR | MAF::NFE2 | 165 | 175 | + | 14.72 | ATGACTGAGCA |
| ERV3-16A3_LTR | ZNF610 | 19 | 28 | + | 14.54 | GCGCCGCTCG |
| ERV3-16A3_LTR | ZNF766 | 359 | 367 | + | 14.38 | AATAAACCT |
| ERV3-16A3_LTR | INSM1 | 153 | 164 | - | 14.22 | TGGCTGGGGGCT |
| ERV3-16A3_LTR | Hoxa13 | 357 | 364 | + | 14.02 | CCAATAAA |
| ERV3-16A3_LTR | TFAP4::FLI1 | 91 | 104 | - | 13.97 | GGCGCTAACAGCTG |
| ERV3-16A3_LTR | NHLH1 | 57 | 65 | - | 13.78 | CGCAGCTGA |
| ERV3-16A3_LTR | MAFG::NFE2L1 | 165 | 175 | + | 13.58 | ATGACTGAGCA |
| ERV3-16A3_LTR | Bach1::Mafk | 164 | 175 | + | 13.46 | AATGACTGAGCA |
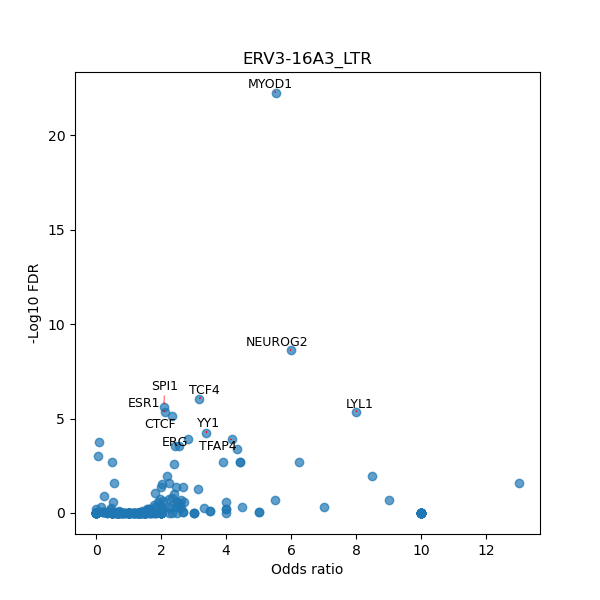
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.