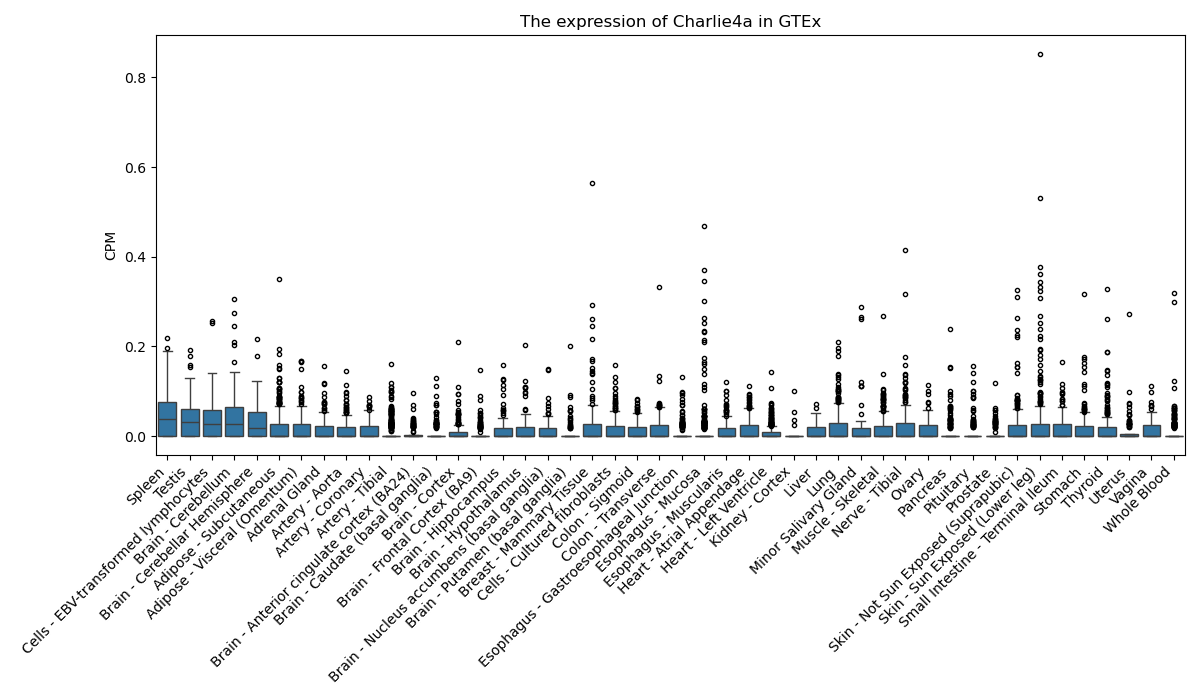
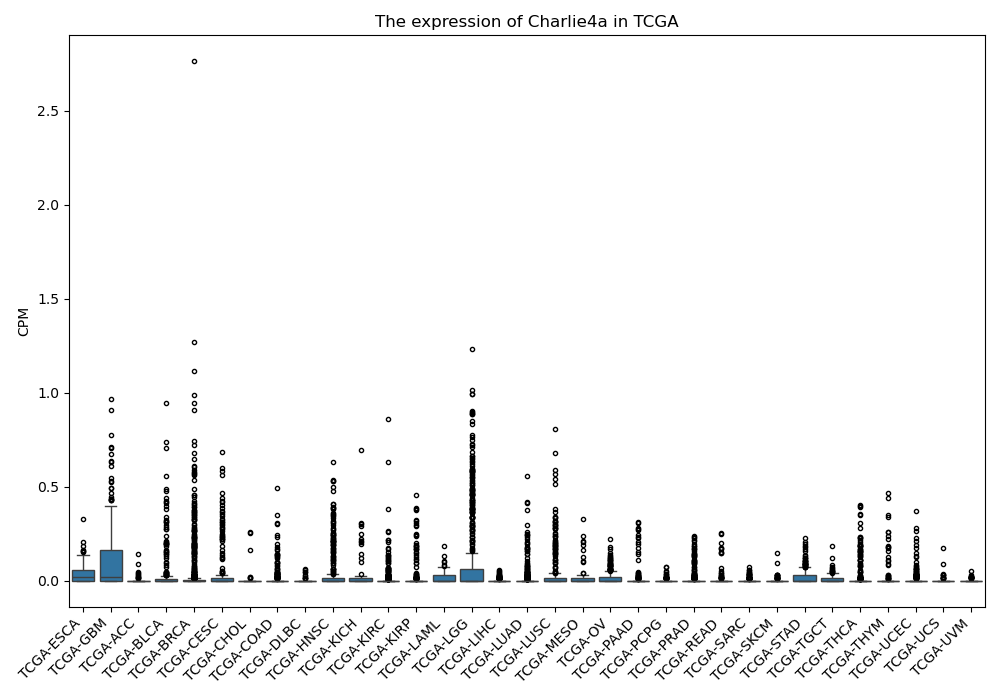
Charlie4a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000025 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 508 |
| Kimura value | 23.45 |
| Tau index | 0.9392 |
| Description | hAT-Charlie DNA transposon, Charlie4a subfamily (non-autonomous) |
| Comment | 16bp TIR, similar to those of the MER1 family. 8bp TSD, with a bias for NTCTAGAN. |
| Sequence |
CAGGGGTTCTTAACCTGGGGTCCGTGGATAGAATTCAGGGGGTCCGTGAACTTGGATGGGGAAAAAAATTACATCTTTATTTTCACTAACCTCTAACTGAAATTTAGCATTTCCTTCAATTATGAATGTAGGCAACAAACCACAGTAGTATTAGCAGTACCTGTGACTTTGTCACCAATAGAAATCACAGATATTTTCATATCACATTACAGTTGTTGCAGATATCTCGAAATATCATTTACGCTCATCACTACTTCGAAATTACGGTAGTTATTAGACCCGCCGCTAGATCTTGTTATTTAATGCGTTAATAAAGAAGCACATATATTACTATATCACAAATTTGNTTTTTTAATATTTTGATAACTGTATTTCAATATAATTGGTTTCCTTTGTAATCCTATGTATTTTATTTTATGCATTTAAAAACATTATTCTGAGAAGGGGTCCATAGGCTTCACCAGACTGCCAAAGGGGTCCATGGCACAAAAAAGGTTAAGAACCCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie4a | Stat5a::Stat5b | 435 | 443 | - | 12.96 | TTCTCAGAA |
| Charlie4a | NR1H4::RXRA | 40 | 52 | - | 12.92 | AGTTCACGGACCC |
| Charlie4a | SATB1 | 271 | 277 | - | 12.89 | CTAATAA |
| Charlie4a | F10B5.3 | 223 | 233 | - | 12.85 | ATTTCGAGATA |
| Charlie4a | hb | 484 | 493 | + | 12.79 | GCACAAAAAA |
| Charlie4a | MYB56 | 363 | 372 | + | 12.58 | ATAACTGTAT |
| Charlie4a | Stat4 | 435 | 444 | + | 12.55 | TTCTGAGAAG |
| Charlie4a | Dr | 380 | 386 | - | 12.51 | CCAATTA |
| Charlie4a | FOXD3 | 74 | 87 | - | 12.50 | AGTGAAAATAAAGA |
| Charlie4a | TB1 | 37 | 45 | - | 12.46 | GGACCCCCT |
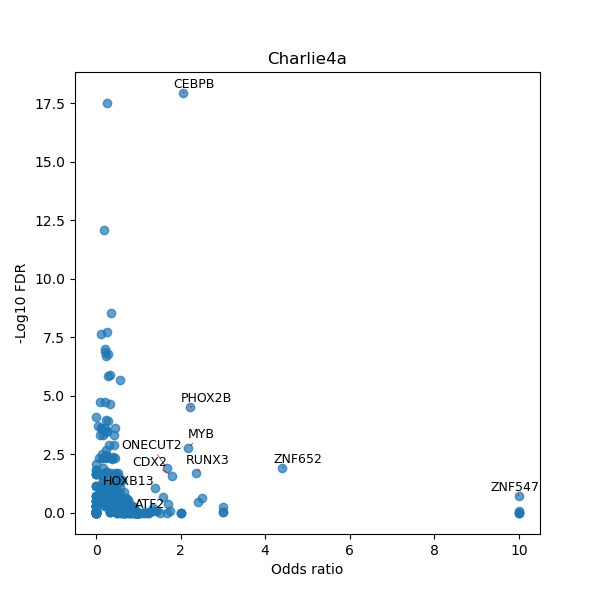
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.