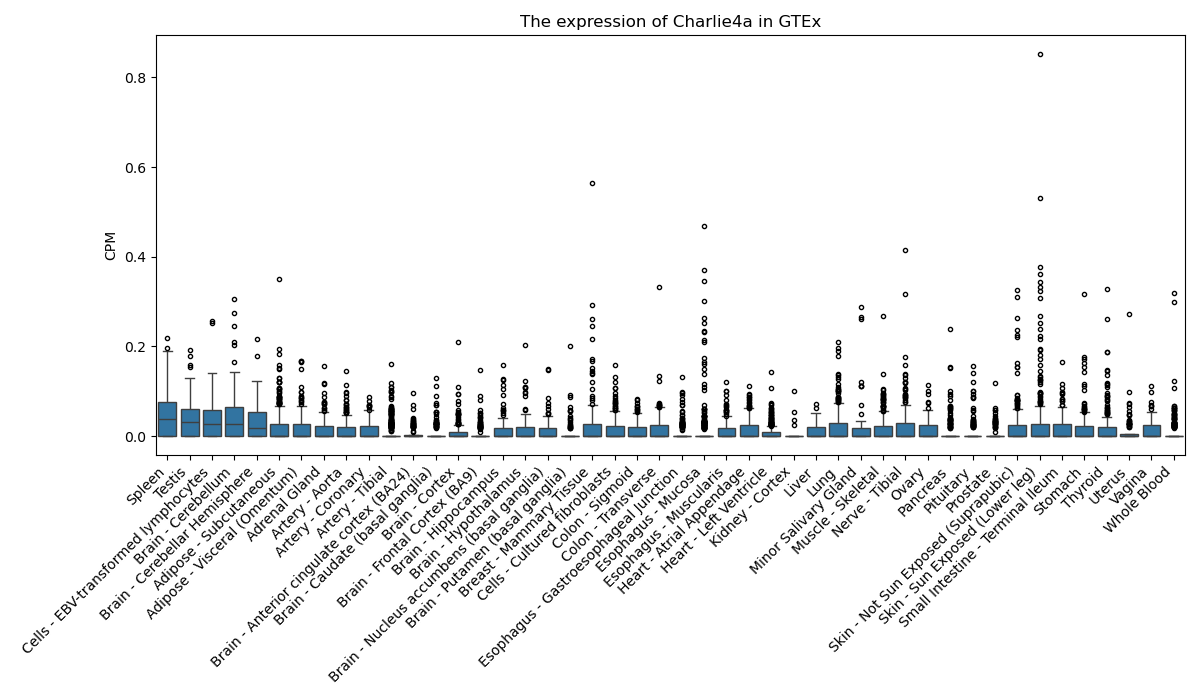
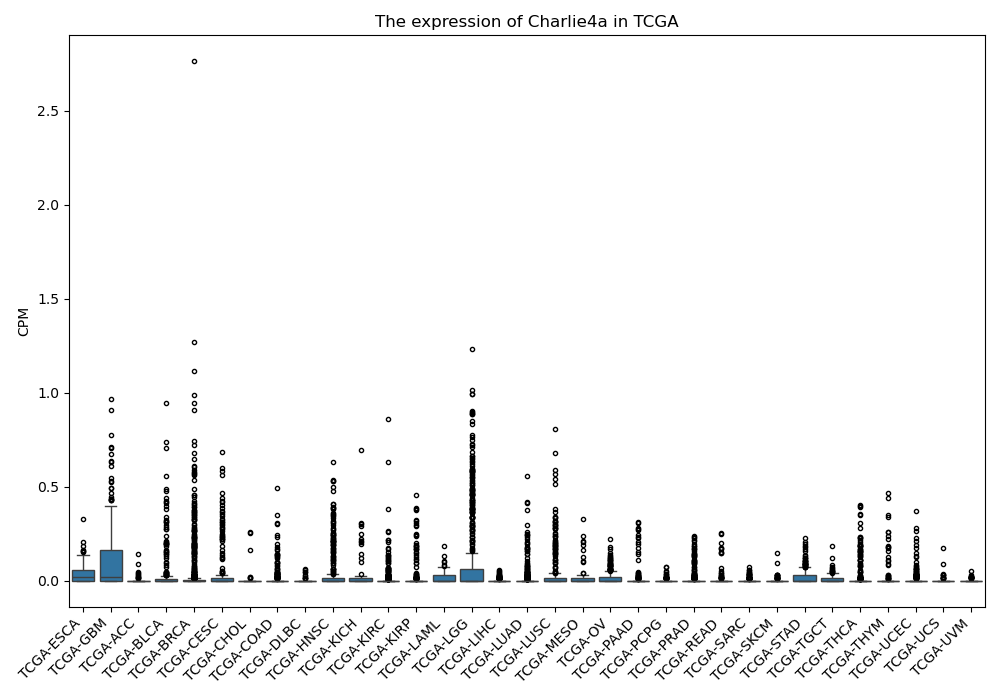
Charlie4a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000025 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 508 |
| Kimura value | 23.45 |
| Tau index | 0.9392 |
| Description | hAT-Charlie DNA transposon, Charlie4a subfamily (non-autonomous) |
| Comment | 16bp TIR, similar to those of the MER1 family. 8bp TSD, with a bias for NTCTAGAN. |
| Sequence |
CAGGGGTTCTTAACCTGGGGTCCGTGGATAGAATTCAGGGGGTCCGTGAACTTGGATGGGGAAAAAAATTACATCTTTATTTTCACTAACCTCTAACTGAAATTTAGCATTTCCTTCAATTATGAATGTAGGCAACAAACCACAGTAGTATTAGCAGTACCTGTGACTTTGTCACCAATAGAAATCACAGATATTTTCATATCACATTACAGTTGTTGCAGATATCTCGAAATATCATTTACGCTCATCACTACTTCGAAATTACGGTAGTTATTAGACCCGCCGCTAGATCTTGTTATTTAATGCGTTAATAAAGAAGCACATATATTACTATATCACAAATTTGNTTTTTTAATATTTTGATAACTGTATTTCAATATAATTGGTTTCCTTTGTAATCCTATGTATTTTATTTTATGCATTTAAAAACATTATTCTGAGAAGGGGTCCATAGGCTTCACCAGACTGCCAAAGGGGTCCATGGCACAAAAAAGGTTAAGAACCCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie4a | RVE4 | 189 | 197 | + | 16.53 | AGATATTTT |
| Charlie4a | RVE7 | 189 | 197 | - | 16.38 | AAAATATCT |
| Charlie4a | RVE6 | 189 | 197 | + | 16.23 | AGATATTTT |
| Charlie4a | RVE5 | 189 | 197 | + | 16.21 | AGATATTTT |
| Charlie4a | RVE8 | 189 | 197 | + | 16.08 | AGATATTTT |
| Charlie4a | USV1 | 34 | 43 | - | 15.98 | ACCCCCTGAA |
| Charlie4a | LHY | 189 | 197 | + | 15.86 | AGATATTTT |
| Charlie4a | RVE1 | 189 | 197 | - | 15.59 | AAAATATCT |
| Charlie4a | ceh-10::ttx-3 | 382 | 395 | + | 15.49 | ATTGGTTTCCTTTG |
| Charlie4a | RVE7L | 189 | 197 | - | 15.47 | AAAATATCT |
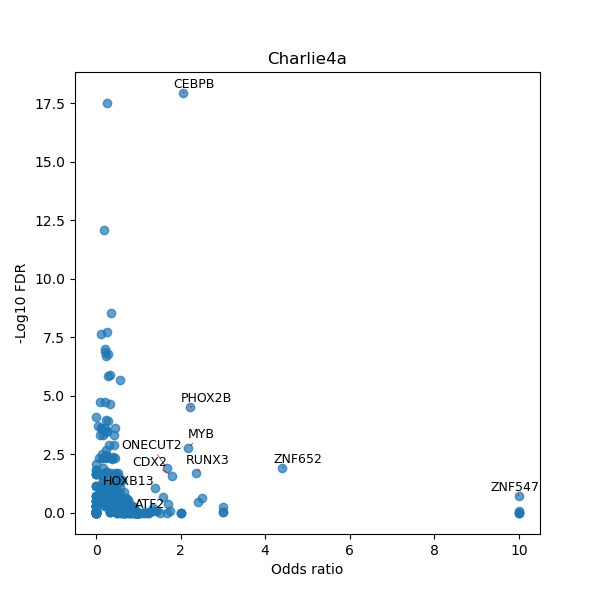
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.