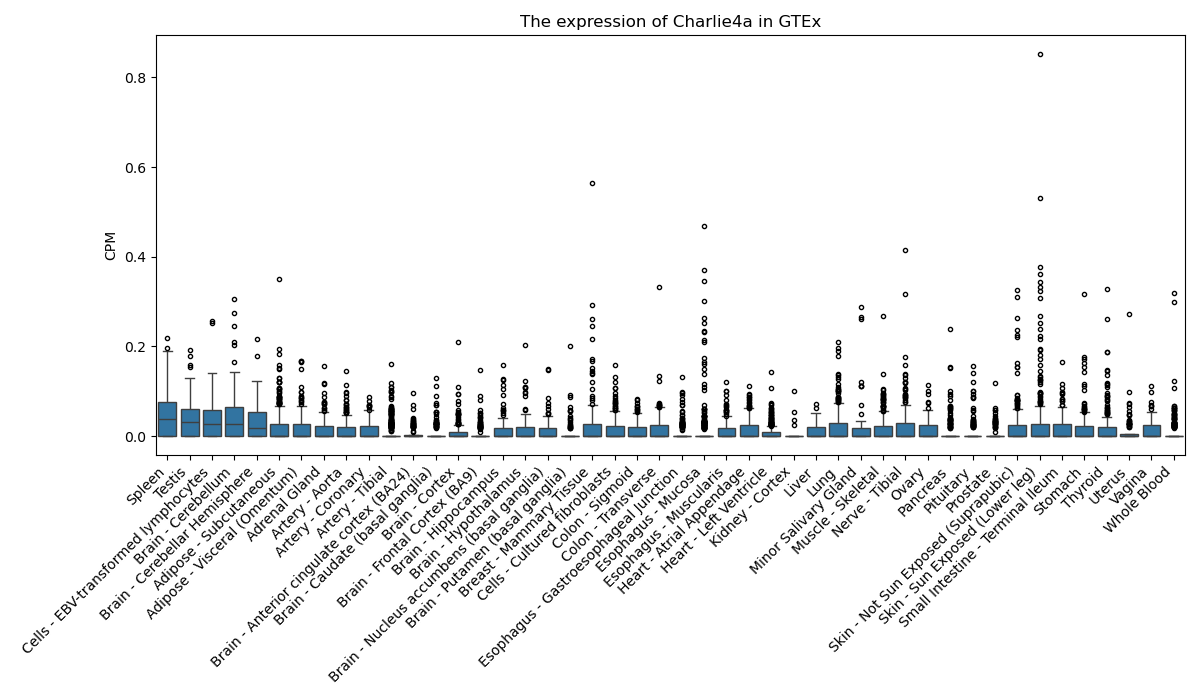
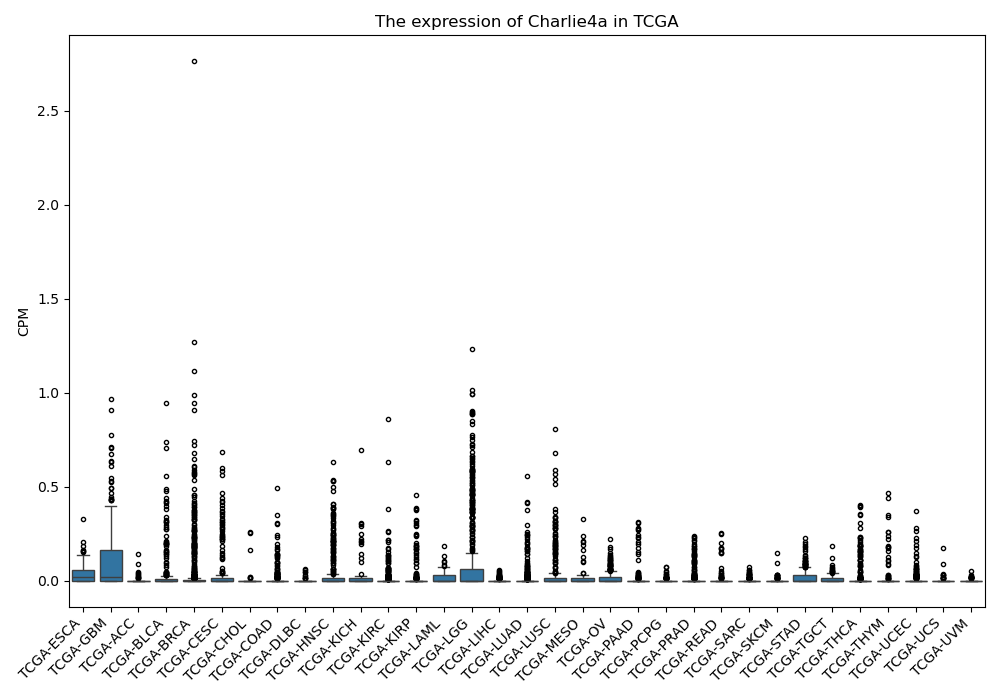
Charlie4a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000025 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 508 |
| Kimura value | 23.45 |
| Tau index | 0.9392 |
| Description | hAT-Charlie DNA transposon, Charlie4a subfamily (non-autonomous) |
| Comment | 16bp TIR, similar to those of the MER1 family. 8bp TSD, with a bias for NTCTAGAN. |
| Sequence |
CAGGGGTTCTTAACCTGGGGTCCGTGGATAGAATTCAGGGGGTCCGTGAACTTGGATGGGGAAAAAAATTACATCTTTATTTTCACTAACCTCTAACTGAAATTTAGCATTTCCTTCAATTATGAATGTAGGCAACAAACCACAGTAGTATTAGCAGTACCTGTGACTTTGTCACCAATAGAAATCACAGATATTTTCATATCACATTACAGTTGTTGCAGATATCTCGAAATATCATTTACGCTCATCACTACTTCGAAATTACGGTAGTTATTAGACCCGCCGCTAGATCTTGTTATTTAATGCGTTAATAAAGAAGCACATATATTACTATATCACAAATTTGNTTTTTTAATATTTTGATAACTGTATTTCAATATAATTGGTTTCCTTTGTAATCCTATGTATTTTATTTTATGCATTTAAAAACATTATTCTGAGAAGGGGTCCATAGGCTTCACCAGACTGCCAAAGGGGTCCATGGCACAAAAAAGGTTAAGAACCCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie4a | ERF118 | 280 | 287 | + | 13.65 | CCGCCGCT |
| Charlie4a | Aef1 | 211 | 218 | - | 13.61 | CAACAACT |
| Charlie4a | D1 | 410 | 417 | - | 13.51 | TAAAATAA |
| Charlie4a | sens | 182 | 191 | + | 13.43 | AAATCACAGA |
| Charlie4a | REI1 | 35 | 41 | - | 13.40 | CCCCTGA |
| Charlie4a | SEP3 | 486 | 495 | - | 13.39 | CCTTTTTTGT |
| Charlie4a | D1 | 296 | 303 | - | 13.38 | TTAAATAA |
| Charlie4a | br | 291 | 302 | - | 13.26 | TAAATAACAAGA |
| Charlie4a | unc-86 | 415 | 422 | + | 13.03 | TTATGCAT |
| Charlie4a | Gfi1B | 182 | 191 | + | 12.98 | AAATCACAGA |
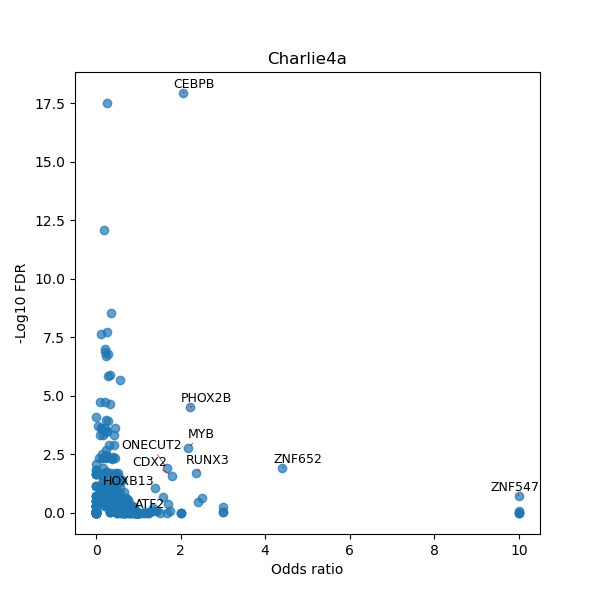
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.