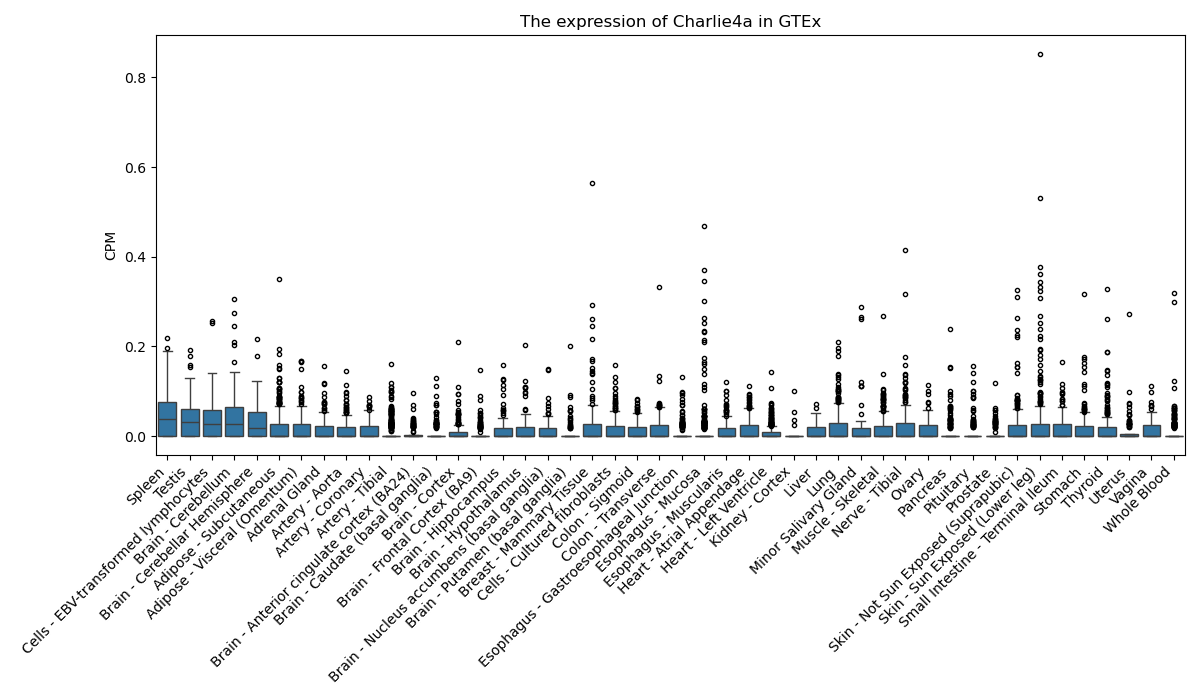
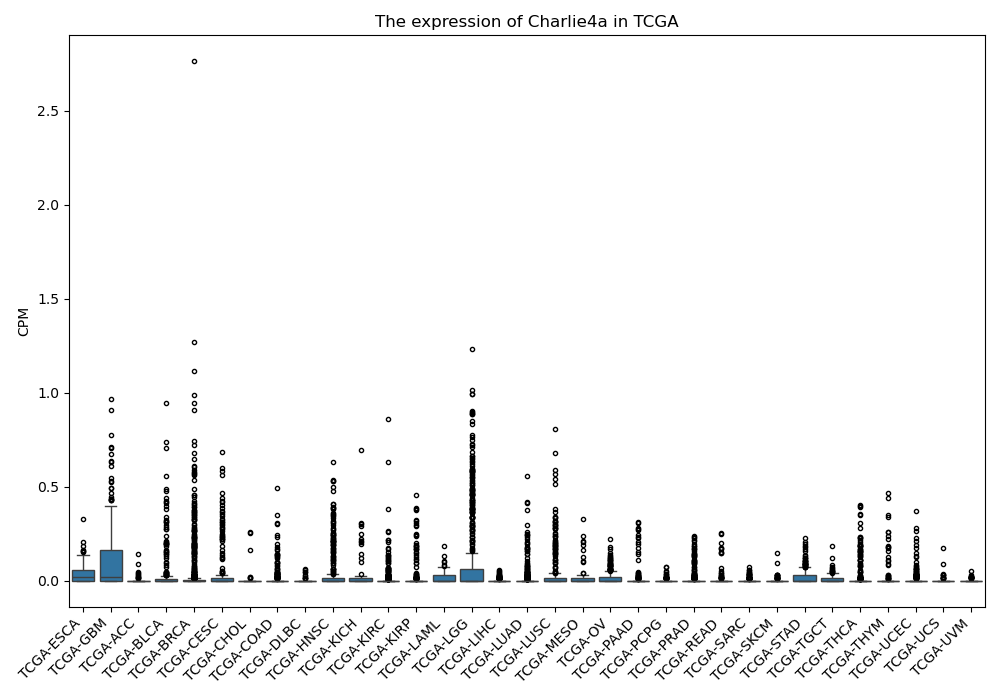
Charlie4a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000025 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 508 |
| Kimura value | 23.45 |
| Tau index | 0.9392 |
| Description | hAT-Charlie DNA transposon, Charlie4a subfamily (non-autonomous) |
| Comment | 16bp TIR, similar to those of the MER1 family. 8bp TSD, with a bias for NTCTAGAN. |
| Sequence |
CAGGGGTTCTTAACCTGGGGTCCGTGGATAGAATTCAGGGGGTCCGTGAACTTGGATGGGGAAAAAAATTACATCTTTATTTTCACTAACCTCTAACTGAAATTTAGCATTTCCTTCAATTATGAATGTAGGCAACAAACCACAGTAGTATTAGCAGTACCTGTGACTTTGTCACCAATAGAAATCACAGATATTTTCATATCACATTACAGTTGTTGCAGATATCTCGAAATATCATTTACGCTCATCACTACTTCGAAATTACGGTAGTTATTAGACCCGCCGCTAGATCTTGTTATTTAATGCGTTAATAAAGAAGCACATATATTACTATATCACAAATTTGNTTTTTTAATATTTTGATAACTGTATTTCAATATAATTGGTTTCCTTTGTAATCCTATGTATTTTATTTTATGCATTTAAAAACATTATTCTGAGAAGGGGTCCATAGGCTTCACCAGACTGCCAAAGGGGTCCATGGCACAAAAAAGGTTAAGAACCCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie4a | DOF4.2 | 489 | 499 | + | 15.27 | AAAAAGGTTAA |
| Charlie4a | ANL2 | 297 | 306 | - | 15.23 | GCATTAAATA |
| Charlie4a | HDG7 | 297 | 306 | - | 15.23 | GCATTAAATA |
| Charlie4a | HDG1 | 297 | 306 | + | 14.99 | TATTTAATGC |
| Charlie4a | CCA1 | 189 | 196 | - | 14.72 | AAATATCT |
| Charlie4a | Runx1 | 137 | 145 | - | 14.41 | CTGTGGTTT |
| Charlie4a | PK02532.1 | 189 | 196 | - | 14.19 | AAATATCT |
| Charlie4a | lsl-1 | 260 | 270 | - | 13.97 | CTACCGTAATT |
| Charlie4a | Stat5b | 435 | 443 | - | 13.85 | TTCTCAGAA |
| Charlie4a | Bcl11B | 137 | 145 | + | 13.67 | AAACCACAG |
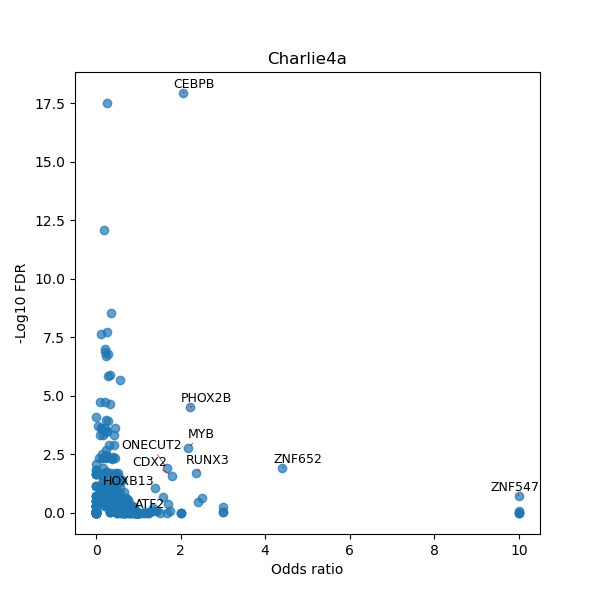
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.