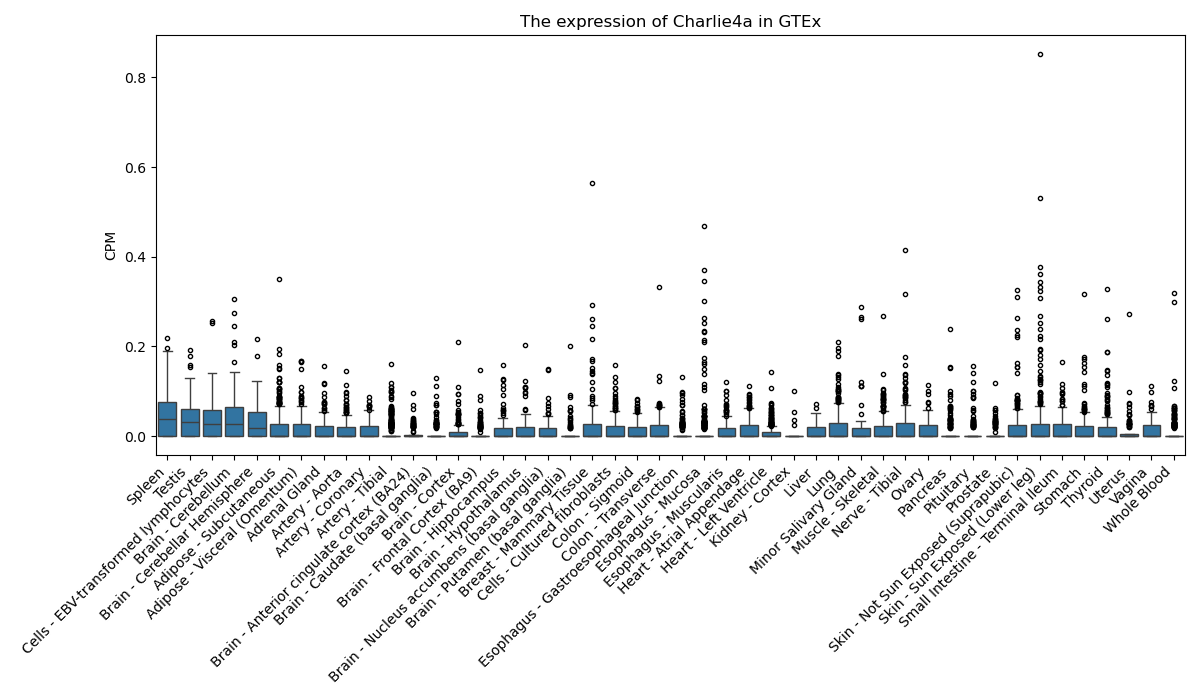
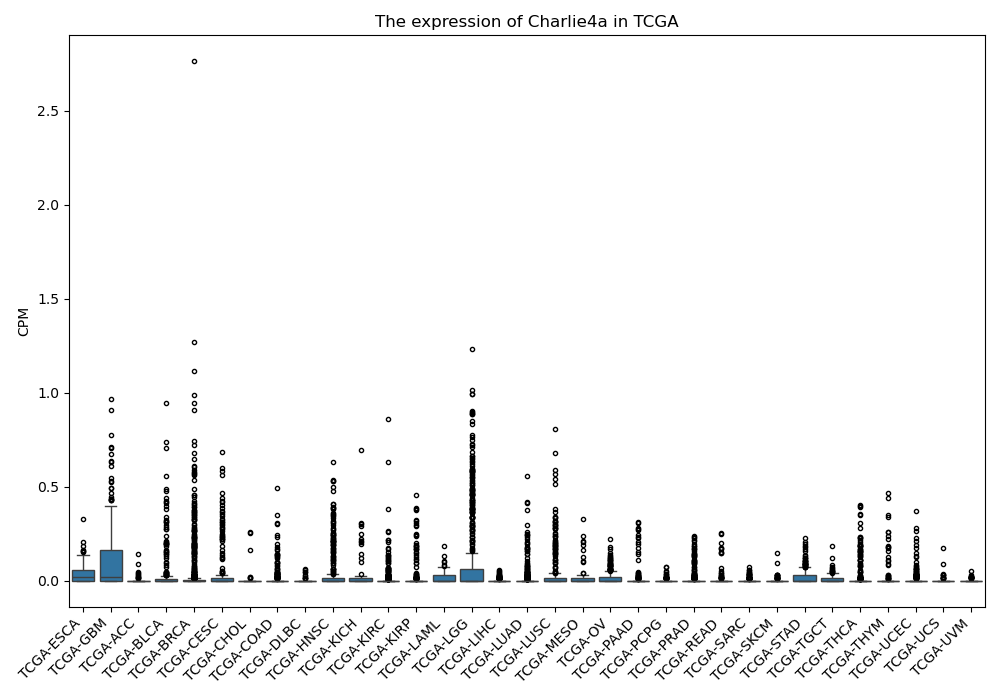
Charlie4a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000025 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 508 |
| Kimura value | 23.45 |
| Tau index | 0.9392 |
| Description | hAT-Charlie DNA transposon, Charlie4a subfamily (non-autonomous) |
| Comment | 16bp TIR, similar to those of the MER1 family. 8bp TSD, with a bias for NTCTAGAN. |
| Sequence |
CAGGGGTTCTTAACCTGGGGTCCGTGGATAGAATTCAGGGGGTCCGTGAACTTGGATGGGGAAAAAAATTACATCTTTATTTTCACTAACCTCTAACTGAAATTTAGCATTTCCTTCAATTATGAATGTAGGCAACAAACCACAGTAGTATTAGCAGTACCTGTGACTTTGTCACCAATAGAAATCACAGATATTTTCATATCACATTACAGTTGTTGCAGATATCTCGAAATATCATTTACGCTCATCACTACTTCGAAATTACGGTAGTTATTAGACCCGCCGCTAGATCTTGTTATTTAATGCGTTAATAAAGAAGCACATATATTACTATATCACAAATTTGNTTTTTTAATATTTTGATAACTGTATTTCAATATAATTGGTTTCCTTTGTAATCCTATGTATTTTATTTTATGCATTTAAAAACATTATTCTGAGAAGGGGTCCATAGGCTTCACCAGACTGCCAAAGGGGTCCATGGCACAAAAAAGGTTAAGAACCCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie4a | HSFA6A | 224 | 236 | - | 4.92 | GATATTTCGAGAT |
| Charlie4a | NR2F6 | 492 | 506 | + | 4.90 | AAGGTTAAGAACCCC |
| Charlie4a | NR2F6 | 492 | 506 | - | 4.71 | GGGGTTCTTAACCTT |
| Charlie4a | Sox1 | 373 | 387 | - | 3.78 | ACCAATTATATTGAA |
| Charlie4a | ZNF558 | 390 | 418 | - | 3.48 | ATAAAATAAAATACATAGGATTACAAAGG |
| Charlie4a | Zm00001d020595 | 278 | 287 | - | 2.65 | AGCGGCGGGT |
| Charlie4a | Sox1 | 373 | 387 | + | 1.76 | TTCAATATAATTGGT |
| Charlie4a | SOX21 | 373 | 387 | - | 0.65 | ACCAATTATATTGAA |
| Charlie4a | ZNF558 | 354 | 382 | - | 0.30 | TTATATTGAAATACAGTTATCAAAATATT |
| Charlie4a | SOX21 | 203 | 217 | - | 0.29 | AACAACTGTAATGTG |
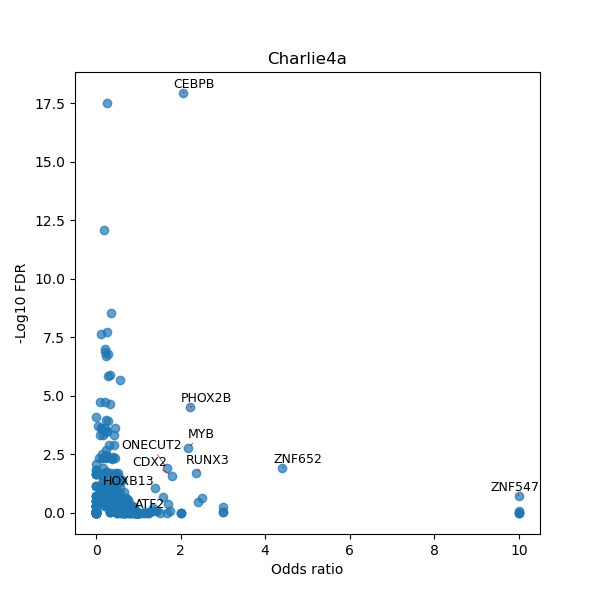
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.