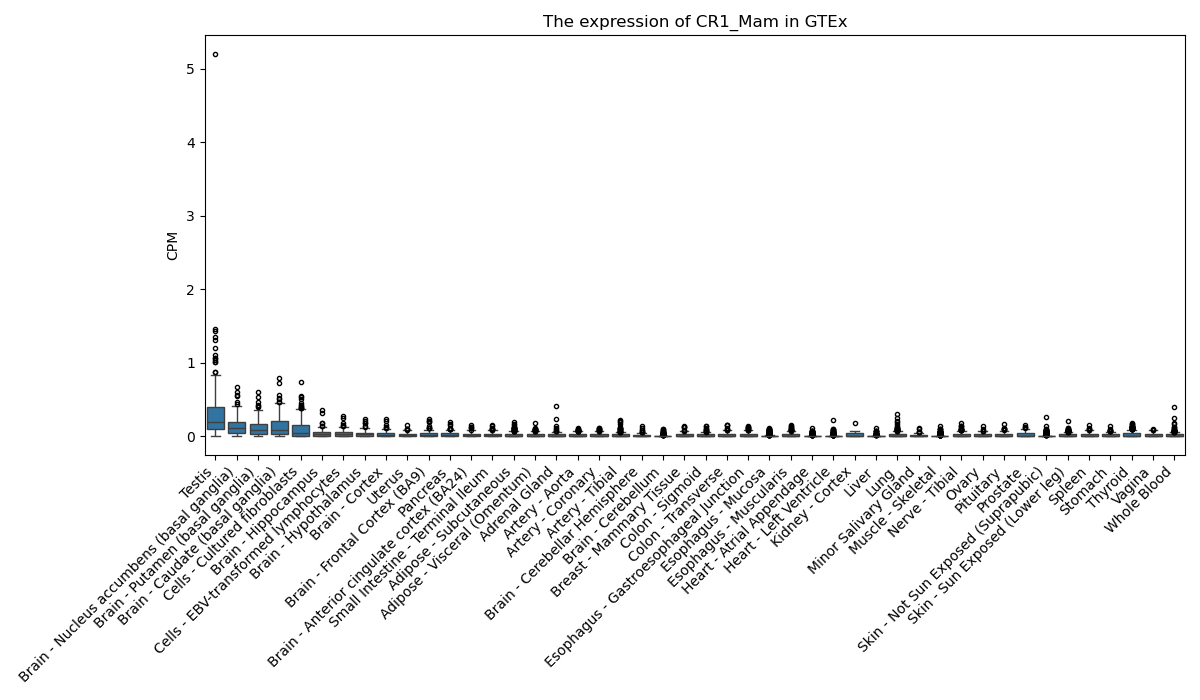
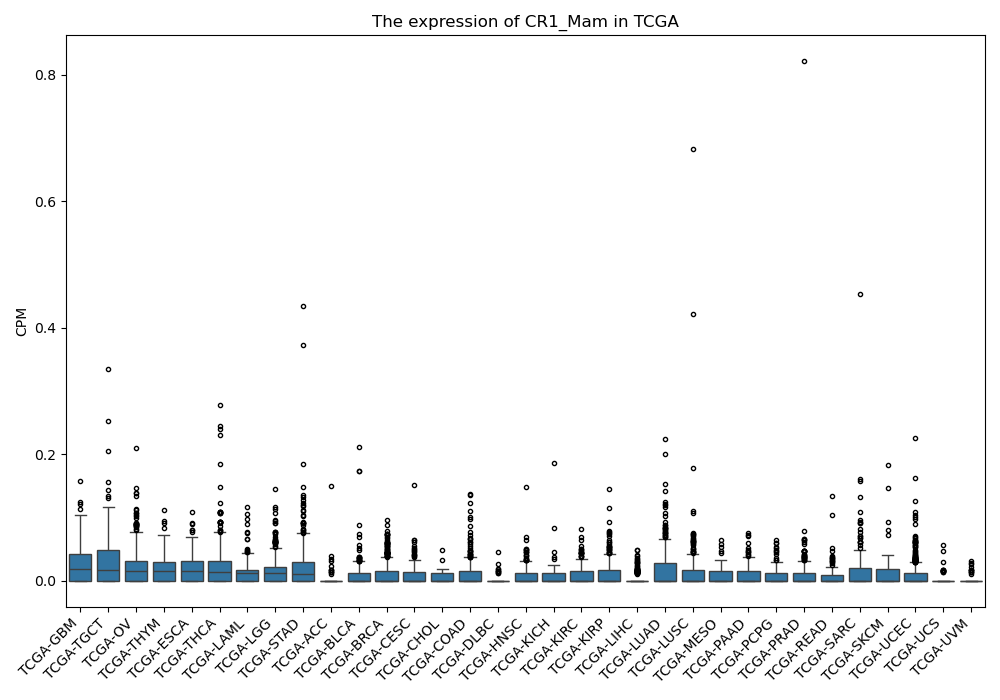
CR1_Mam
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000110 |
|---|---|
| TE superfamily | CR1 |
| TE class | LINE |
| Species | Mammalia |
| Length | 2204 |
| Kimura value | 35.29 |
| Tau index | 0.9420 |
| Description | Mammalian CR1 (Chicken Repeat 1) LINE |
| Comment | Mammalian remnant of an ancient LINE element first identified as CR1 (Chicken Repeat 1). The vast majority of CR1_Mam corresponds to roughly 2/3rds of ORF2 from CR1. |
| Sequence |
TAGATTGGCAAAATACCTCATCGGGACGAAATATAGAAGAGANGCTTTTCGATAAGACAAATNACCGCTTCCTAGAATAGGTAGTTTTAGAACCCACAAGGAAAGGAAATANACTGGACTTGGTTCTGAGNAGTGAACAGGAACTGGCGCAGGAAGTATCTGTGGGTGAACCACTATGTGACCAATAACGATTAAATTCAACATTCTGGCGGGAAAGGAAAACCAAAAAAAATCCATCACATGTTACTCAATTTTAGAAAAGGGAACTATGACAAAATGTGGGCATTAGTTAGGAAAAATGAAAAGNANAGTTTAAAAAGTAAATAACCCACAGGAAGCCTGGAGACTATTTNAAAACACCTTATTGGAAGCCCAGGAAAAATGTGTGCCANGGATCAAAAANACCAGGCGCTTGGAAAAAAAAATACCCNGCATGGCTAACTGGCAAAGTCAAAGAGGCTATTGTAGGGGAAAAGGCATCTTTCAAAAAATGGAAAGANAANTCCAAGTGAGGAGAACAGGGAAGCCCGCAAAGTATGGCAGGTCAGGTACAAACTAGAAATTAGGCGGGCCAAAAAAGAATTTGAGGAGCACCTTGCAAGGGACTCCAAAGCCAATAATAAAAGGTTCTTTAAATACATCAAAGGNAGGAAGCCAGCTAGAGAATCAGTGGGACCATTTGATGATCACGGTGCAAAAGGTGNACTCAAGATGAAAAAGGAGATAGCAGAGAGACTNAATGAATTCTTTGCTTCGGTCTTTACTGAGGAAGACGTTAGGGAGATTCCCACTCCCAAATTGTCTTTTGAAGCAGACAGGTCAGAGGTACTAGATCAAATAGTGGTAAACGCACAGGATGTTCTAGACCTAATTGACAAACTAGATGTAGATAAATCACCGGACCAGATGGCATCCACCCAAGAGTTTTGAAGGAACTCAAGGGTGAAACTGTTGACAAAATGTGTAACCTGTATTACAAACGGCCACTGTGCCNAGAACNAGTGTGTTCAATGGGCTCCATCCATAAGAAGGGCTCCAGAGGCCATGGAACTAAAGCCAGTGAGTCTCACTTCCATACCNGGCAAGCTGGTGGNATATAAGCAAAGAGTAGNACTNANCGCGCANNCAAACATAACCTACTAGGAGAAAGGCAACATGGTTTCTGTAAGGGGAAATCATGCCTGACTAATCTATTAGAGTTCTTTGAGGGGGTAAATAACATGTGGACAAGGAGAAACCAGTGGATATAATTTATTTAGACTTTCAAAAAGCCTTTGACAAGGTTCCACACCAAAACTTTATTTTTAAAAATTGAGTCACCATGGGATTNGGGGGAATGTTTTGTCATGGATAGGGAACTGGCTTAGAGACAGGAAACAAAAGGTAGAGAATAAACGGGCACTTCTCTGGATGGAGAAGTGTAAACAGTGGGGTCCCCCAGGGATCAGTGCTGGGACTAGTCTTATTTAACATTTTTATAAATGATCTGGAAGAGGGAGTGCACAGTGAAATCTCCAAGTTTGCAGATGACACTAAGCTCTTCCGGGTAGTGAAATGCCAAGCCGATGGGGATAAACTGCAGGAAGATCTCACGAGGCTGTGTGAGTGGGCAGAAAAGTGGCAGATGAGCTTCAGTGTGGGCAAGTGTAAGGTAATGCATTTAGGGAAAAATAATCCAAACTATACTTATAAGATGATGGGCTCTGAGCTATCAGTTACGACCCAGGAAAGGGATCTAGGAGTCATTGTAGACTGTTCCCTGAAGACATCAGCCCAATGTGCTGTAGCCAAAAAGGCCAACAAAATGCTGGGCATCATCAGGAAGGGTATTGGAAACAAAACAGAAAACATTATCCTACCTTTGTACAAAACCATGGTGTATCCGCACCTGGAATACTGTGTGCAGTTCTGGTCGCCGCATCCCTCAAGAAAGACATAACAGAGCTGAGAAGGGTAATTAAAATGATCAAGGGGATGGAAGGCTTTNATAAGAGGCTGAGCTGNTTTGTTTAAAACTTTAGTCTGGAAAGACGAAGGCTGAGAGGGGATATGATCAAAGTCTATAAAATCATGAAGGGTATGGATAAGGTGAACACGGACTTGTTCACCAAATCCCAGAATACTAGAACTAGGGCANCCTTTGAAGCTTGAAAGAAATTTTAGGGACAAATAAAAGAAAAGGCGTANACTCATTCTACAGAGGG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| CR1_Mam | EWSR1-FLI1 | 1727 | 1744 | + | -15.74 | GAAAGGGATCTAGGAGTC |
| CR1_Mam | EWSR1-FLI1 | 140 | 157 | + | -15.93 | GGAACTGGCGCAGGAAGT |
| CR1_Mam | BPC5 | 1366 | 1395 | + | -41.34 | AGAGACAGGAAACAAAAGGTAGAGAATAAA |
| CR1_Mam | BPC5 | 649 | 678 | + | -44.44 | AGGAAGCCAGCTAGAGAATCAGTGGGACCA |
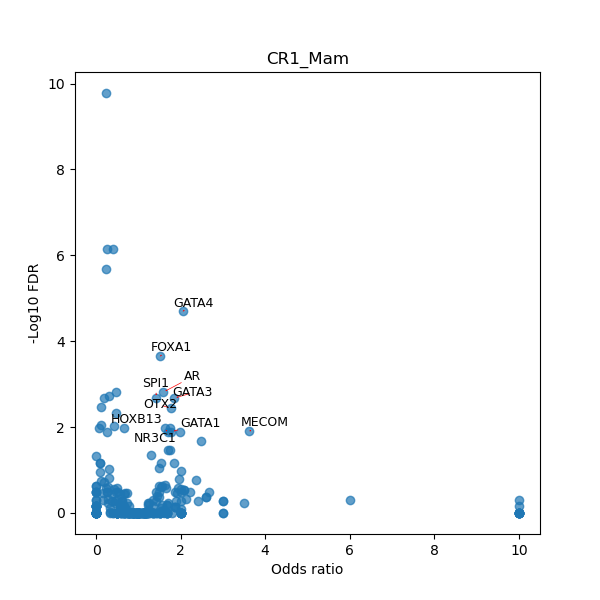
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.