CR1_Mam
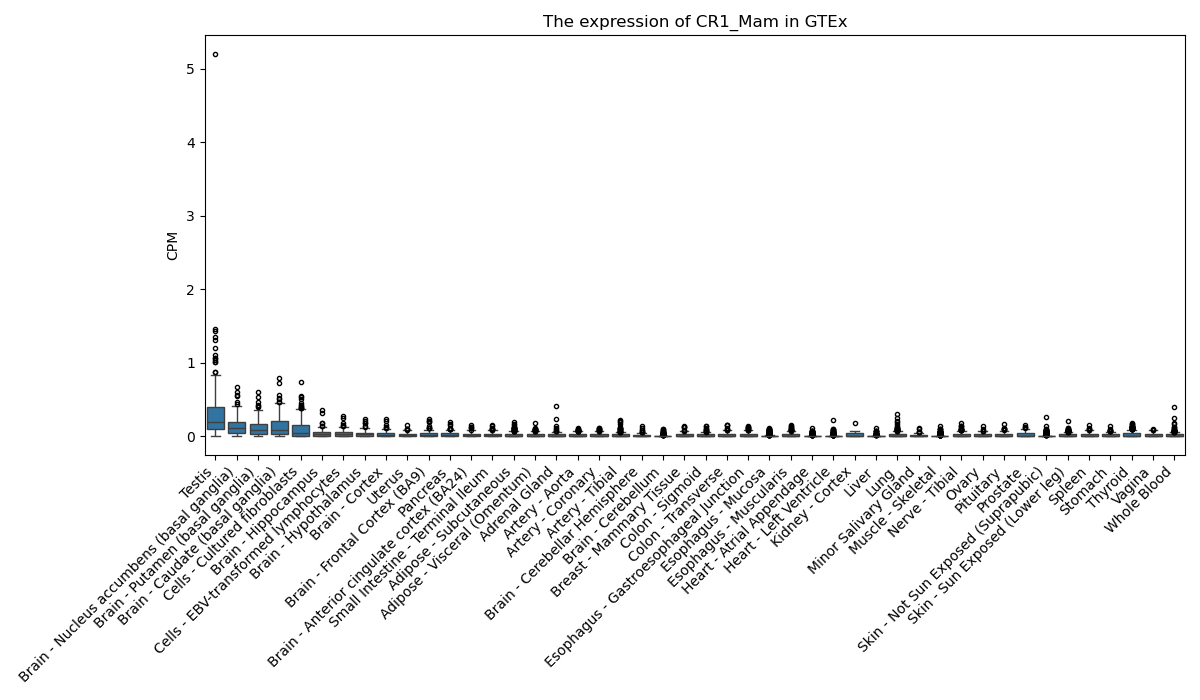
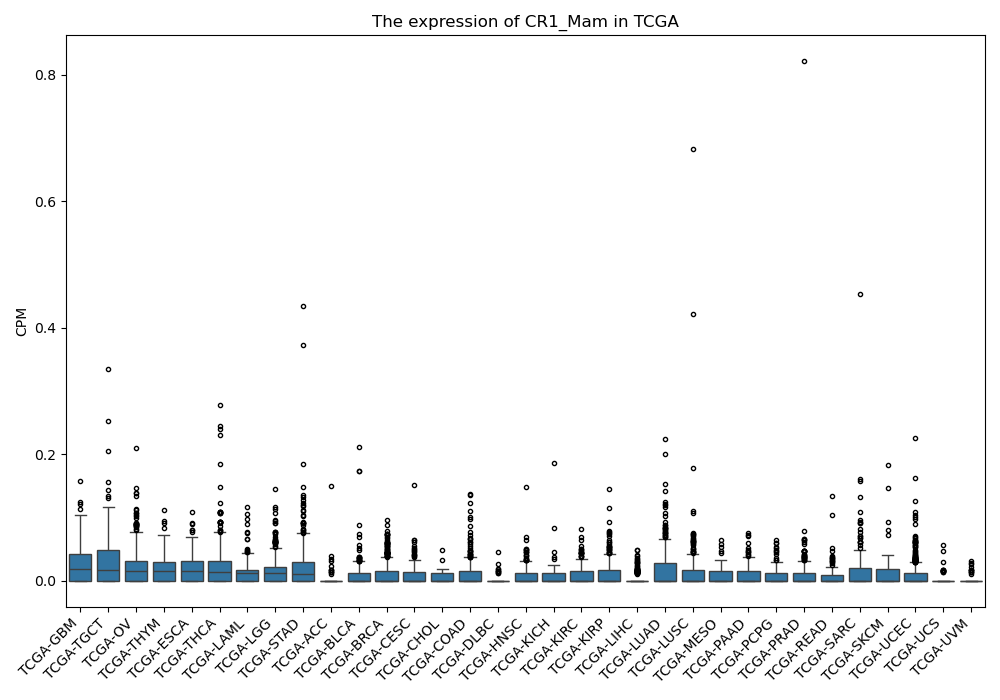
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000110 |
|---|---|
| TE superfamily | CR1 |
| TE class | LINE |
| Species | Mammalia |
| Length | 2204 |
| Kimura value | 35.29 |
| Tau index | 0.9420 |
| Description | Mammalian CR1 (Chicken Repeat 1) LINE |
| Comment | Mammalian remnant of an ancient LINE element first identified as CR1 (Chicken Repeat 1). The vast majority of CR1_Mam corresponds to roughly 2/3rds of ORF2 from CR1. |
| Sequence |
TAGATTGGCAAAATACCTCATCGGGACGAAATATAGAAGAGANGCTTTTCGATAAGACAAATNACCGCTTCCTAGAATAGGTAGTTTTAGAACCCACAAGGAAAGGAAATANACTGGACTTGGTTCTGAGNAGTGAACAGGAACTGGCGCAGGAAGTATCTGTGGGTGAACCACTATGTGACCAATAACGATTAAATTCAACATTCTGGCGGGAAAGGAAAACCAAAAAAAATCCATCACATGTTACTCAATTTTAGAAAAGGGAACTATGACAAAATGTGGGCATTAGTTAGGAAAAATGAAAAGNANAGTTTAAAAAGTAAATAACCCACAGGAAGCCTGGAGACTATTTNAAAACACCTTATTGGAAGCCCAGGAAAAATGTGTGCCANGGATCAAAAANACCAGGCGCTTGGAAAAAAAAATACCCNGCATGGCTAACTGGCAAAGTCAAAGAGGCTATTGTAGGGGAAAAGGCATCTTTCAAAAAATGGAAAGANAANTCCAAGTGAGGAGAACAGGGAAGCCCGCAAAGTATGGCAGGTCAGGTACAAACTAGAAATTAGGCGGGCCAAAAAAGAATTTGAGGAGCACCTTGCAAGGGACTCCAAAGCCAATAATAAAAGGTTCTTTAAATACATCAAAGGNAGGAAGCCAGCTAGAGAATCAGTGGGACCATTTGATGATCACGGTGCAAAAGGTGNACTCAAGATGAAAAAGGAGATAGCAGAGAGACTNAATGAATTCTTTGCTTCGGTCTTTACTGAGGAAGACGTTAGGGAGATTCCCACTCCCAAATTGTCTTTTGAAGCAGACAGGTCAGAGGTACTAGATCAAATAGTGGTAAACGCACAGGATGTTCTAGACCTAATTGACAAACTAGATGTAGATAAATCACCGGACCAGATGGCATCCACCCAAGAGTTTTGAAGGAACTCAAGGGTGAAACTGTTGACAAAATGTGTAACCTGTATTACAAACGGCCACTGTGCCNAGAACNAGTGTGTTCAATGGGCTCCATCCATAAGAAGGGCTCCAGAGGCCATGGAACTAAAGCCAGTGAGTCTCACTTCCATACCNGGCAAGCTGGTGGNATATAAGCAAAGAGTAGNACTNANCGCGCANNCAAACATAACCTACTAGGAGAAAGGCAACATGGTTTCTGTAAGGGGAAATCATGCCTGACTAATCTATTAGAGTTCTTTGAGGGGGTAAATAACATGTGGACAAGGAGAAACCAGTGGATATAATTTATTTAGACTTTCAAAAAGCCTTTGACAAGGTTCCACACCAAAACTTTATTTTTAAAAATTGAGTCACCATGGGATTNGGGGGAATGTTTTGTCATGGATAGGGAACTGGCTTAGAGACAGGAAACAAAAGGTAGAGAATAAACGGGCACTTCTCTGGATGGAGAAGTGTAAACAGTGGGGTCCCCCAGGGATCAGTGCTGGGACTAGTCTTATTTAACATTTTTATAAATGATCTGGAAGAGGGAGTGCACAGTGAAATCTCCAAGTTTGCAGATGACACTAAGCTCTTCCGGGTAGTGAAATGCCAAGCCGATGGGGATAAACTGCAGGAAGATCTCACGAGGCTGTGTGAGTGGGCAGAAAAGTGGCAGATGAGCTTCAGTGTGGGCAAGTGTAAGGTAATGCATTTAGGGAAAAATAATCCAAACTATACTTATAAGATGATGGGCTCTGAGCTATCAGTTACGACCCAGGAAAGGGATCTAGGAGTCATTGTAGACTGTTCCCTGAAGACATCAGCCCAATGTGCTGTAGCCAAAAAGGCCAACAAAATGCTGGGCATCATCAGGAAGGGTATTGGAAACAAAACAGAAAACATTATCCTACCTTTGTACAAAACCATGGTGTATCCGCACCTGGAATACTGTGTGCAGTTCTGGTCGCCGCATCCCTCAAGAAAGACATAACAGAGCTGAGAAGGGTAATTAAAATGATCAAGGGGATGGAAGGCTTTNATAAGAGGCTGAGCTGNTTTGTTTAAAACTTTAGTCTGGAAAGACGAAGGCTGAGAGGGGATATGATCAAAGTCTATAAAATCATGAAGGGTATGGATAAGGTGAACACGGACTTGTTCACCAAATCCCAGAATACTAGAACTAGGGCANCCTTTGAAGCTTGAAAGAAATTTTAGGGACAAATAAAAGAAAAGGCGTANACTCATTCTACAGAGGG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| CR1_Mam | SVP | 482 | 497 | + | 18.05 | TTTCAAAAAATGGAAA |
| CR1_Mam | DOF4.3 | 622 | 634 | - | 17.18 | TAAAGAACCTTTT |
| CR1_Mam | AGL63 | 483 | 497 | + | 17.11 | TTCAAAAAATGGAAA |
| CR1_Mam | AT1G19000 | 2078 | 2090 | + | 16.97 | GTATGGATAAGGT |
| CR1_Mam | FLC | 485 | 498 | + | 16.40 | CAAAAAATGGAAAG |
| CR1_Mam | PI | 485 | 498 | + | 16.35 | CAAAAAATGGAAAG |
| CR1_Mam | ZIM3 | 1839 | 1849 | + | 16.05 | AAACAGAAAAC |
| CR1_Mam | nhr-33 | 548 | 560 | + | 16.02 | GGTACAAACTAGA |
| CR1_Mam | WRKY46 | 447 | 456 | - | 16.02 | CTTTGACTTT |
| CR1_Mam | ZKSCAN5 | 1066 | 1074 | - | 15.83 | GGAAGTGAG |
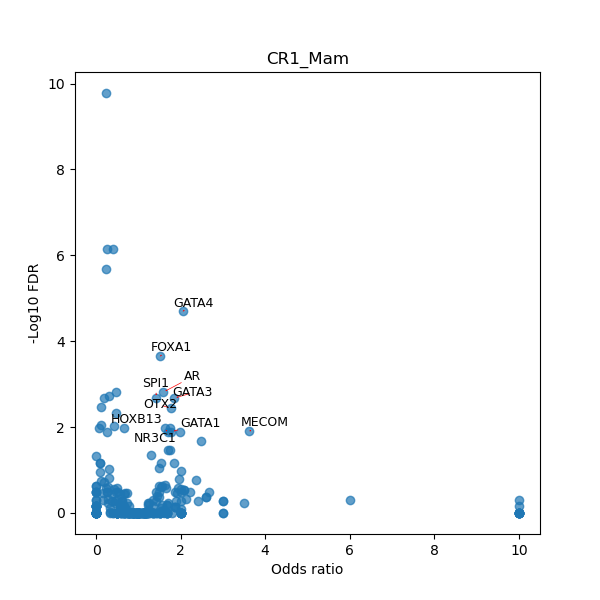
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.