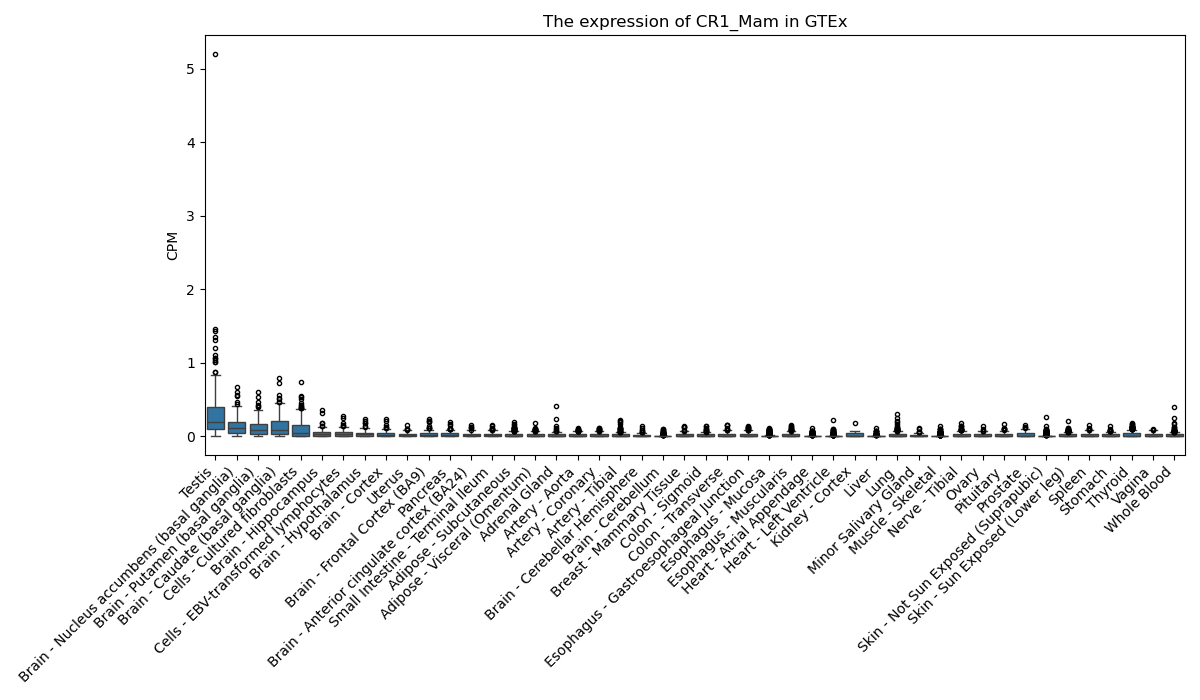
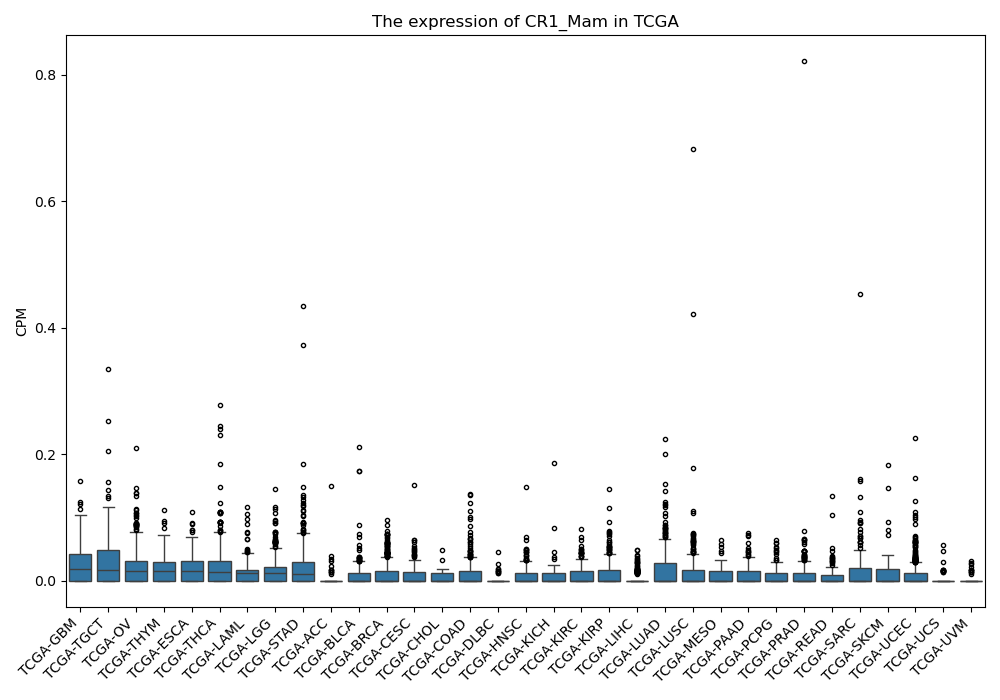
CR1_Mam
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000110 |
|---|---|
| TE superfamily | CR1 |
| TE class | LINE |
| Species | Mammalia |
| Length | 2204 |
| Kimura value | 35.29 |
| Tau index | 0.9420 |
| Description | Mammalian CR1 (Chicken Repeat 1) LINE |
| Comment | Mammalian remnant of an ancient LINE element first identified as CR1 (Chicken Repeat 1). The vast majority of CR1_Mam corresponds to roughly 2/3rds of ORF2 from CR1. |
| Sequence |
TAGATTGGCAAAATACCTCATCGGGACGAAATATAGAAGAGANGCTTTTCGATAAGACAAATNACCGCTTCCTAGAATAGGTAGTTTTAGAACCCACAAGGAAAGGAAATANACTGGACTTGGTTCTGAGNAGTGAACAGGAACTGGCGCAGGAAGTATCTGTGGGTGAACCACTATGTGACCAATAACGATTAAATTCAACATTCTGGCGGGAAAGGAAAACCAAAAAAAATCCATCACATGTTACTCAATTTTAGAAAAGGGAACTATGACAAAATGTGGGCATTAGTTAGGAAAAATGAAAAGNANAGTTTAAAAAGTAAATAACCCACAGGAAGCCTGGAGACTATTTNAAAACACCTTATTGGAAGCCCAGGAAAAATGTGTGCCANGGATCAAAAANACCAGGCGCTTGGAAAAAAAAATACCCNGCATGGCTAACTGGCAAAGTCAAAGAGGCTATTGTAGGGGAAAAGGCATCTTTCAAAAAATGGAAAGANAANTCCAAGTGAGGAGAACAGGGAAGCCCGCAAAGTATGGCAGGTCAGGTACAAACTAGAAATTAGGCGGGCCAAAAAAGAATTTGAGGAGCACCTTGCAAGGGACTCCAAAGCCAATAATAAAAGGTTCTTTAAATACATCAAAGGNAGGAAGCCAGCTAGAGAATCAGTGGGACCATTTGATGATCACGGTGCAAAAGGTGNACTCAAGATGAAAAAGGAGATAGCAGAGAGACTNAATGAATTCTTTGCTTCGGTCTTTACTGAGGAAGACGTTAGGGAGATTCCCACTCCCAAATTGTCTTTTGAAGCAGACAGGTCAGAGGTACTAGATCAAATAGTGGTAAACGCACAGGATGTTCTAGACCTAATTGACAAACTAGATGTAGATAAATCACCGGACCAGATGGCATCCACCCAAGAGTTTTGAAGGAACTCAAGGGTGAAACTGTTGACAAAATGTGTAACCTGTATTACAAACGGCCACTGTGCCNAGAACNAGTGTGTTCAATGGGCTCCATCCATAAGAAGGGCTCCAGAGGCCATGGAACTAAAGCCAGTGAGTCTCACTTCCATACCNGGCAAGCTGGTGGNATATAAGCAAAGAGTAGNACTNANCGCGCANNCAAACATAACCTACTAGGAGAAAGGCAACATGGTTTCTGTAAGGGGAAATCATGCCTGACTAATCTATTAGAGTTCTTTGAGGGGGTAAATAACATGTGGACAAGGAGAAACCAGTGGATATAATTTATTTAGACTTTCAAAAAGCCTTTGACAAGGTTCCACACCAAAACTTTATTTTTAAAAATTGAGTCACCATGGGATTNGGGGGAATGTTTTGTCATGGATAGGGAACTGGCTTAGAGACAGGAAACAAAAGGTAGAGAATAAACGGGCACTTCTCTGGATGGAGAAGTGTAAACAGTGGGGTCCCCCAGGGATCAGTGCTGGGACTAGTCTTATTTAACATTTTTATAAATGATCTGGAAGAGGGAGTGCACAGTGAAATCTCCAAGTTTGCAGATGACACTAAGCTCTTCCGGGTAGTGAAATGCCAAGCCGATGGGGATAAACTGCAGGAAGATCTCACGAGGCTGTGTGAGTGGGCAGAAAAGTGGCAGATGAGCTTCAGTGTGGGCAAGTGTAAGGTAATGCATTTAGGGAAAAATAATCCAAACTATACTTATAAGATGATGGGCTCTGAGCTATCAGTTACGACCCAGGAAAGGGATCTAGGAGTCATTGTAGACTGTTCCCTGAAGACATCAGCCCAATGTGCTGTAGCCAAAAAGGCCAACAAAATGCTGGGCATCATCAGGAAGGGTATTGGAAACAAAACAGAAAACATTATCCTACCTTTGTACAAAACCATGGTGTATCCGCACCTGGAATACTGTGTGCAGTTCTGGTCGCCGCATCCCTCAAGAAAGACATAACAGAGCTGAGAAGGGTAATTAAAATGATCAAGGGGATGGAAGGCTTTNATAAGAGGCTGAGCTGNTTTGTTTAAAACTTTAGTCTGGAAAGACGAAGGCTGAGAGGGGATATGATCAAAGTCTATAAAATCATGAAGGGTATGGATAAGGTGAACACGGACTTGTTCACCAAATCCCAGAATACTAGAACTAGGGCANCCTTTGAAGCTTGAAAGAAATTTTAGGGACAAATAAAAGAAAAGGCGTANACTCATTCTACAGAGGG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| CR1_Mam | Ebf2 | 1436 | 1444 | + | 14.72 | CCCCAGGGA |
| CR1_Mam | TFDP1 | 208 | 215 | + | 14.70 | GGCGGGAA |
| CR1_Mam | WRKY45 | 446 | 455 | + | 14.67 | CAAAGTCAAA |
| CR1_Mam | CDF5 | 2166 | 2186 | - | 14.64 | TACGCCTTTTCTTTTATTTGT |
| CR1_Mam | TCF7L1 | 636 | 647 | + | 14.59 | ATACATCAAAGG |
| CR1_Mam | AP1 | 485 | 497 | + | 14.58 | CAAAAAATGGAAA |
| CR1_Mam | WRKY26 | 446 | 456 | + | 14.56 | CAAAGTCAAAG |
| CR1_Mam | Crg-1 | 317 | 327 | + | 14.56 | AAAGTAAATAA |
| CR1_Mam | pha-4 | 1418 | 1427 | + | 14.53 | AGTGTAAACA |
| CR1_Mam | blmp-1 | 295 | 305 | + | 14.51 | AAAAATGAAAA |
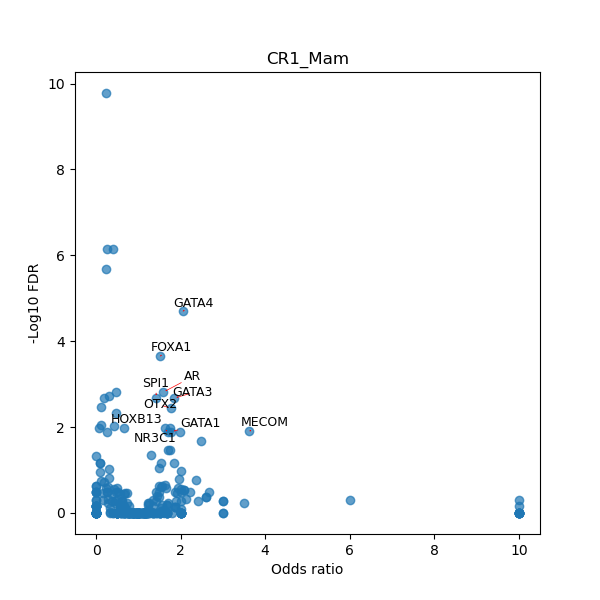
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.