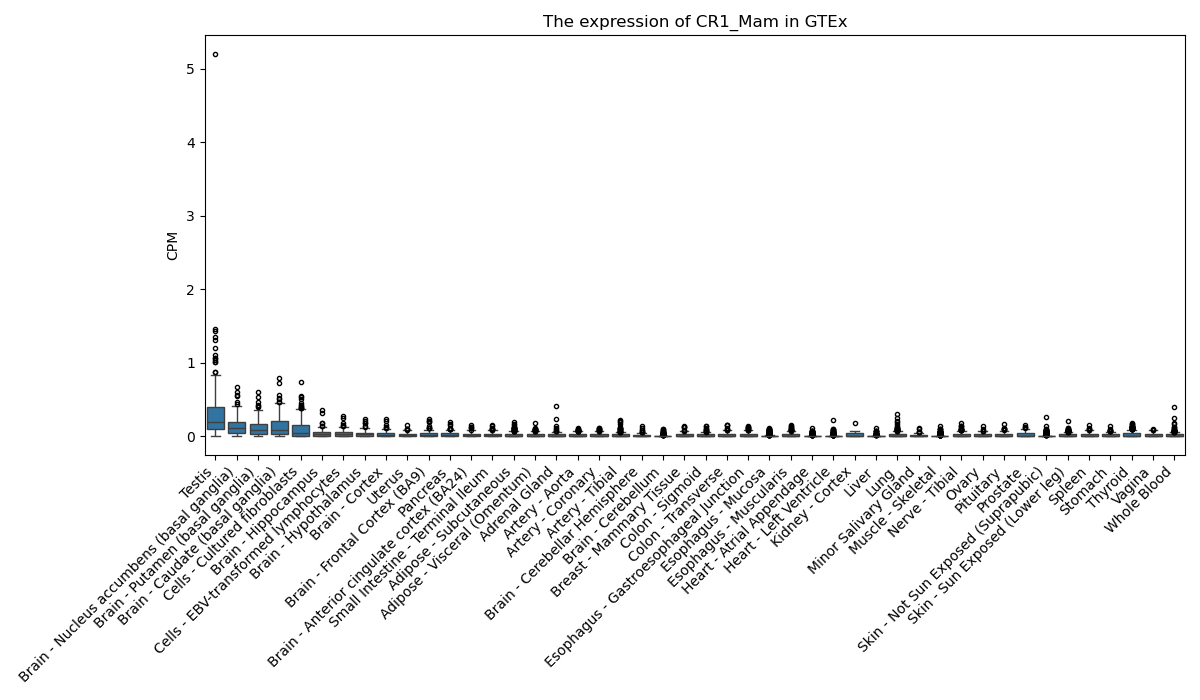
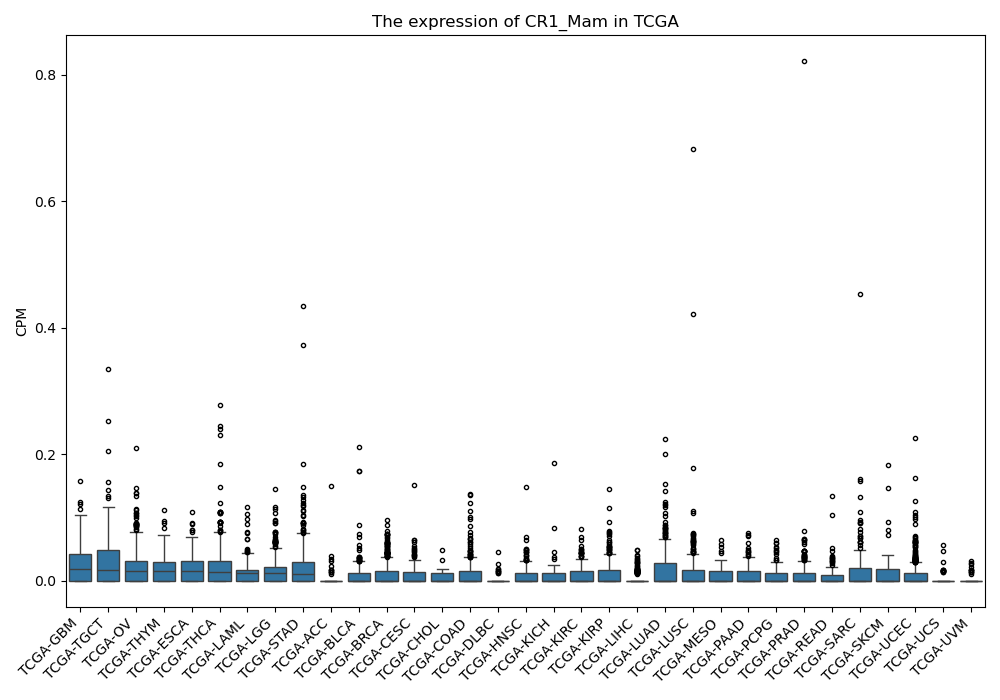
CR1_Mam
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000110 |
|---|---|
| TE superfamily | CR1 |
| TE class | LINE |
| Species | Mammalia |
| Length | 2204 |
| Kimura value | 35.29 |
| Tau index | 0.9420 |
| Description | Mammalian CR1 (Chicken Repeat 1) LINE |
| Comment | Mammalian remnant of an ancient LINE element first identified as CR1 (Chicken Repeat 1). The vast majority of CR1_Mam corresponds to roughly 2/3rds of ORF2 from CR1. |
| Sequence |
TAGATTGGCAAAATACCTCATCGGGACGAAATATAGAAGAGANGCTTTTCGATAAGACAAATNACCGCTTCCTAGAATAGGTAGTTTTAGAACCCACAAGGAAAGGAAATANACTGGACTTGGTTCTGAGNAGTGAACAGGAACTGGCGCAGGAAGTATCTGTGGGTGAACCACTATGTGACCAATAACGATTAAATTCAACATTCTGGCGGGAAAGGAAAACCAAAAAAAATCCATCACATGTTACTCAATTTTAGAAAAGGGAACTATGACAAAATGTGGGCATTAGTTAGGAAAAATGAAAAGNANAGTTTAAAAAGTAAATAACCCACAGGAAGCCTGGAGACTATTTNAAAACACCTTATTGGAAGCCCAGGAAAAATGTGTGCCANGGATCAAAAANACCAGGCGCTTGGAAAAAAAAATACCCNGCATGGCTAACTGGCAAAGTCAAAGAGGCTATTGTAGGGGAAAAGGCATCTTTCAAAAAATGGAAAGANAANTCCAAGTGAGGAGAACAGGGAAGCCCGCAAAGTATGGCAGGTCAGGTACAAACTAGAAATTAGGCGGGCCAAAAAAGAATTTGAGGAGCACCTTGCAAGGGACTCCAAAGCCAATAATAAAAGGTTCTTTAAATACATCAAAGGNAGGAAGCCAGCTAGAGAATCAGTGGGACCATTTGATGATCACGGTGCAAAAGGTGNACTCAAGATGAAAAAGGAGATAGCAGAGAGACTNAATGAATTCTTTGCTTCGGTCTTTACTGAGGAAGACGTTAGGGAGATTCCCACTCCCAAATTGTCTTTTGAAGCAGACAGGTCAGAGGTACTAGATCAAATAGTGGTAAACGCACAGGATGTTCTAGACCTAATTGACAAACTAGATGTAGATAAATCACCGGACCAGATGGCATCCACCCAAGAGTTTTGAAGGAACTCAAGGGTGAAACTGTTGACAAAATGTGTAACCTGTATTACAAACGGCCACTGTGCCNAGAACNAGTGTGTTCAATGGGCTCCATCCATAAGAAGGGCTCCAGAGGCCATGGAACTAAAGCCAGTGAGTCTCACTTCCATACCNGGCAAGCTGGTGGNATATAAGCAAAGAGTAGNACTNANCGCGCANNCAAACATAACCTACTAGGAGAAAGGCAACATGGTTTCTGTAAGGGGAAATCATGCCTGACTAATCTATTAGAGTTCTTTGAGGGGGTAAATAACATGTGGACAAGGAGAAACCAGTGGATATAATTTATTTAGACTTTCAAAAAGCCTTTGACAAGGTTCCACACCAAAACTTTATTTTTAAAAATTGAGTCACCATGGGATTNGGGGGAATGTTTTGTCATGGATAGGGAACTGGCTTAGAGACAGGAAACAAAAGGTAGAGAATAAACGGGCACTTCTCTGGATGGAGAAGTGTAAACAGTGGGGTCCCCCAGGGATCAGTGCTGGGACTAGTCTTATTTAACATTTTTATAAATGATCTGGAAGAGGGAGTGCACAGTGAAATCTCCAAGTTTGCAGATGACACTAAGCTCTTCCGGGTAGTGAAATGCCAAGCCGATGGGGATAAACTGCAGGAAGATCTCACGAGGCTGTGTGAGTGGGCAGAAAAGTGGCAGATGAGCTTCAGTGTGGGCAAGTGTAAGGTAATGCATTTAGGGAAAAATAATCCAAACTATACTTATAAGATGATGGGCTCTGAGCTATCAGTTACGACCCAGGAAAGGGATCTAGGAGTCATTGTAGACTGTTCCCTGAAGACATCAGCCCAATGTGCTGTAGCCAAAAAGGCCAACAAAATGCTGGGCATCATCAGGAAGGGTATTGGAAACAAAACAGAAAACATTATCCTACCTTTGTACAAAACCATGGTGTATCCGCACCTGGAATACTGTGTGCAGTTCTGGTCGCCGCATCCCTCAAGAAAGACATAACAGAGCTGAGAAGGGTAATTAAAATGATCAAGGGGATGGAAGGCTTTNATAAGAGGCTGAGCTGNTTTGTTTAAAACTTTAGTCTGGAAAGACGAAGGCTGAGAGGGGATATGATCAAAGTCTATAAAATCATGAAGGGTATGGATAAGGTGAACACGGACTTGTTCACCAAATCCCAGAATACTAGAACTAGGGCANCCTTTGAAGCTTGAAAGAAATTTTAGGGACAAATAAAAGAAAAGGCGTANACTCATTCTACAGAGGG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| CR1_Mam | tll | 446 | 455 | + | 15.04 | CAAAGTCAAA |
| CR1_Mam | DOF4.2 | 1790 | 1800 | + | 14.97 | AAAAAGGCCAA |
| CR1_Mam | STAT1 | 69 | 77 | - | 14.93 | TTCTAGGAA |
| CR1_Mam | TCF7L2 | 638 | 646 | + | 14.93 | ACATCAAAG |
| CR1_Mam | SOC1 | 485 | 497 | - | 14.90 | TTTCCATTTTTTG |
| CR1_Mam | EBF3 | 1436 | 1444 | + | 14.82 | CCCCAGGGA |
| CR1_Mam | SGR5 | 50 | 62 | + | 14.80 | CGATAAGACAAAT |
| CR1_Mam | WRKY21 | 446 | 456 | + | 14.76 | CAAAGTCAAAG |
| CR1_Mam | WRKY22 | 447 | 456 | + | 14.73 | AAAGTCAAAG |
| CR1_Mam | DOT6 | 15 | 24 | + | 14.72 | ACCTCATCGG |
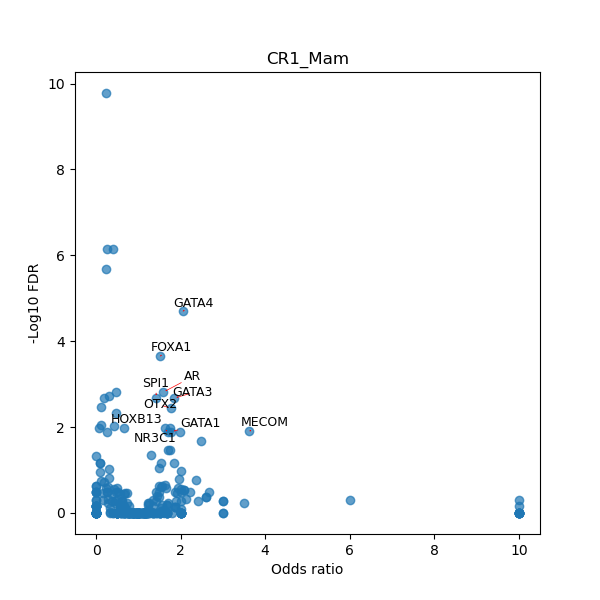
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.