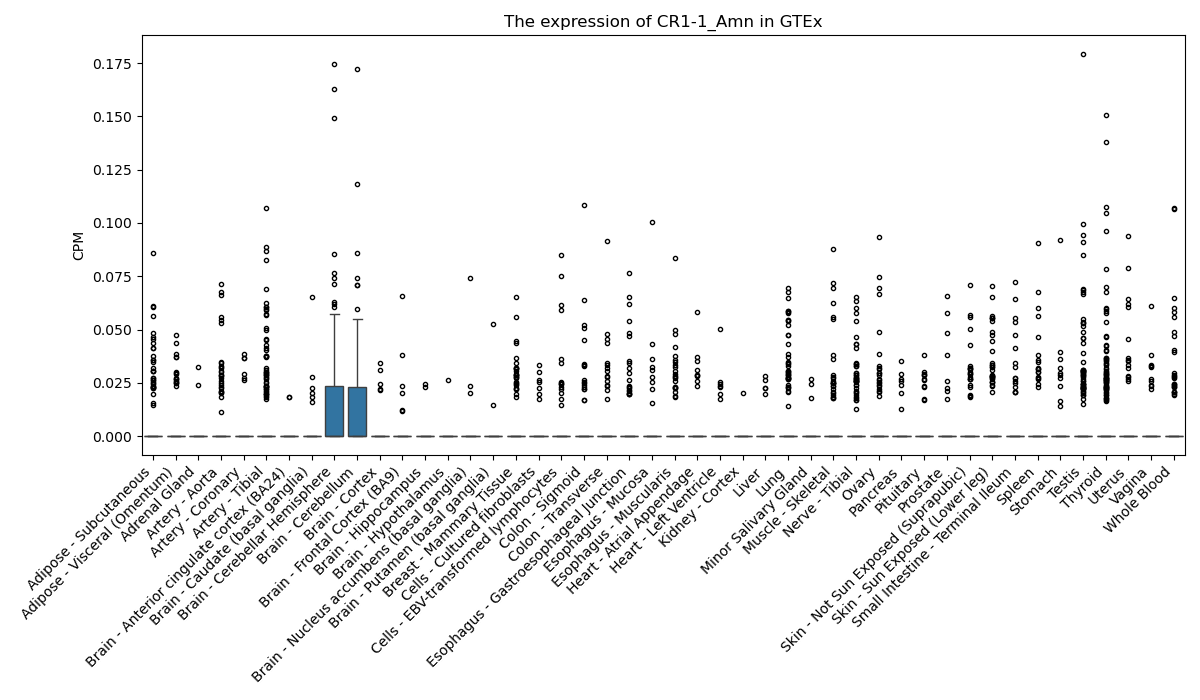
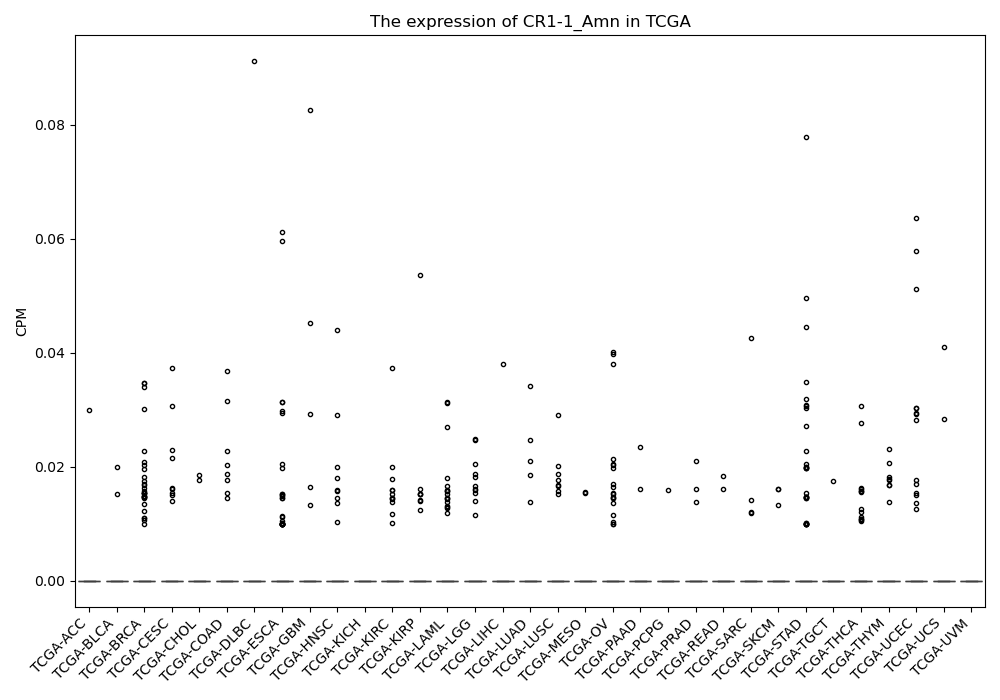
CR1-1_Amn
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001336 |
|---|---|
| TE superfamily | CR1 |
| TE class | LINE |
| Species | Amniota |
| Length | 813 |
| Kimura value | 30.90 |
| Tau index | 0.0000 |
| Description | CR1 (Chicken Repeat 1) retrotransposon, CR1-1_Amn subfamily |
| Comment | - |
| Sequence |
CGAGAAAGGACTTGGGGTNGTGGTGGATAAAACTCTAAAACCTTCGGCACAGTGCTTAACAGCAGTNAAAAAAGCAAACAAAATNCTGGGATGCATAAATAGAGGTATCGAATATAAGTCGGAGGAAATTATTCTAGCACTGTATAAGGGACTAGTGAGGCCACATTTGGAATACTGTGCGCAGTTCTGGTCACCTTATTATAAAAAGGATATTATAGAAATGGAGAAGGTACAGCGGAGGGCAACCAGAATGATACAGGGACTCGGGGATCTTAGTTATTCAGAAAGGTTAGAGAAACTAGGTCTGTATTCTTTGGAAAAGAGGAGATTAAGAGGGGACCTCACTGAGGTATACANAATTCTGAAGGGAATAGAGAGGATAGATGCTACAAGGCTATTTGAGCTAGTGTCAAATACAAGAACAAGGGGGCACGAACTNAAGCTAAGGAAGGGCCGGGCACTAAAGGAATTTAGGCGGAATTTCTTTACGCAGAGNATAGTTAATATATGGAACTCATTGCCACAGGANGTCGTGGAGGCNAGNANTATAACCGGGTTCAAAAGGGGACTGGATAAATTCATGGAGGATAGGTCCATTAGTGGCTATTAAACAGAACGGNTAAGGAATGNTGTCCCTAGCCTCTGNTTGTCGGAGGGTGGAGANGGATGGCAGGAGANAGATCACTTGATCATTACCTGTTAGGTTCACTCCCTCTGAAGCATCTGGTACTGGCCACTGTCGGNAGACAGGATACTGGGCTAGATGGACCNTTGGTCTGACCCAGTATGGCAGTTCTTATGTTCTTATGTTCT
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| CR1-1_Amn | ZNF708 | 228 | 236 | - | 16.48 | GCTGTACCT |
| CR1-1_Amn | pha-4 | 71 | 80 | + | 14.61 | AAAGCAAACA |
| CR1-1_Amn | ROX1 | 418 | 429 | - | 14.31 | CCCCTTGTTCTT |
| CR1-1_Amn | Stat6 | 310 | 319 | - | 14.02 | TTCCAAAGAA |
| CR1-1_Amn | PRDM1 | 2 | 8 | - | 13.28 | CTTTCTC |
| CR1-1_Amn | Nkx2-1 | 683 | 689 | + | 13.15 | CACTTGA |
| CR1-1_Amn | unc-86 | 91 | 98 | - | 13.03 | TTATGCAT |
| CR1-1_Amn | ARF3 | 648 | 655 | + | 12.99 | TGTCGGAG |
| CR1-1_Amn | FOXC2 | 71 | 81 | + | 12.97 | AAAGCAAACAA |
| CR1-1_Amn | LYS14 | 476 | 482 | + | 12.96 | CGGAATT |
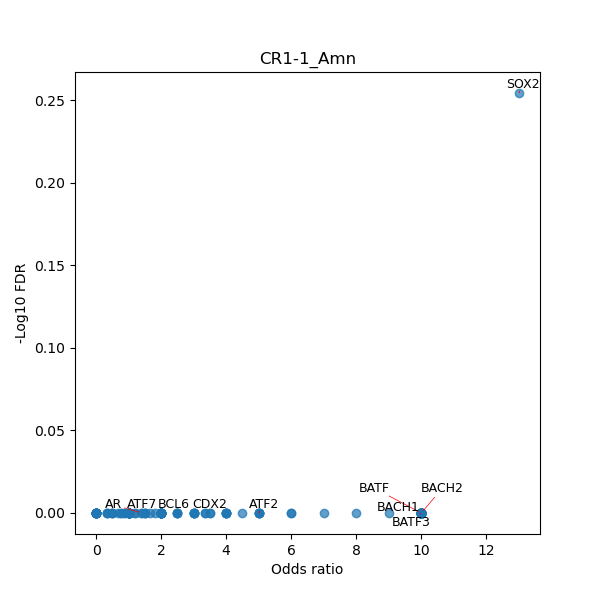
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.