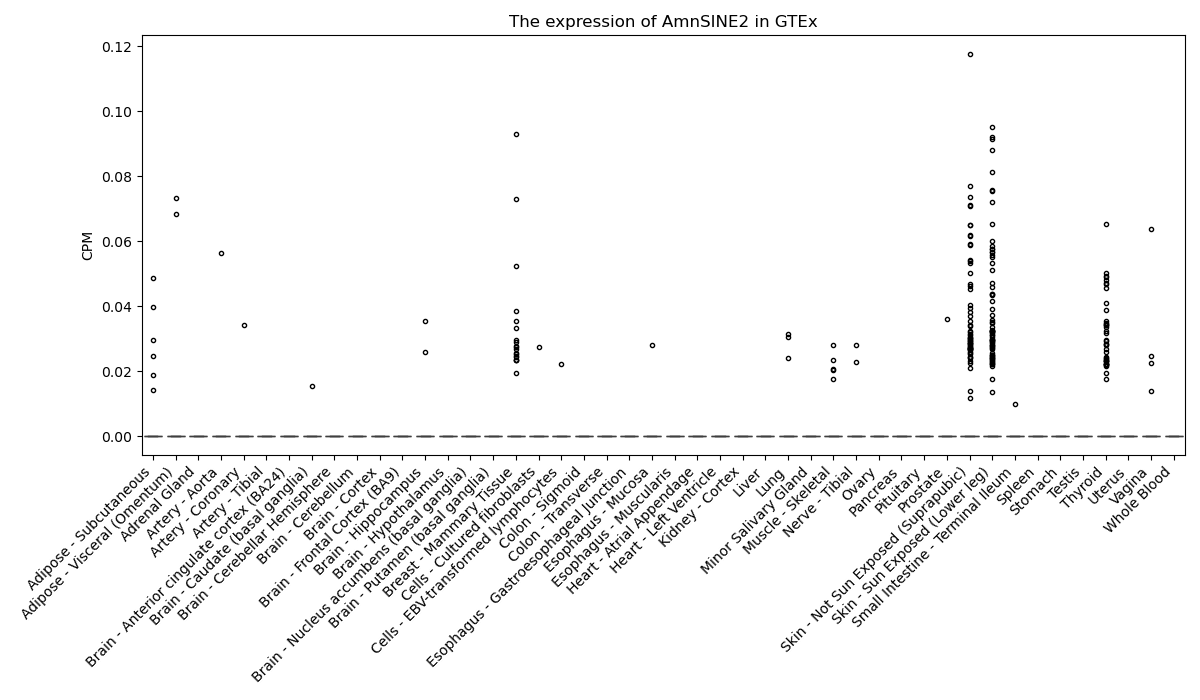
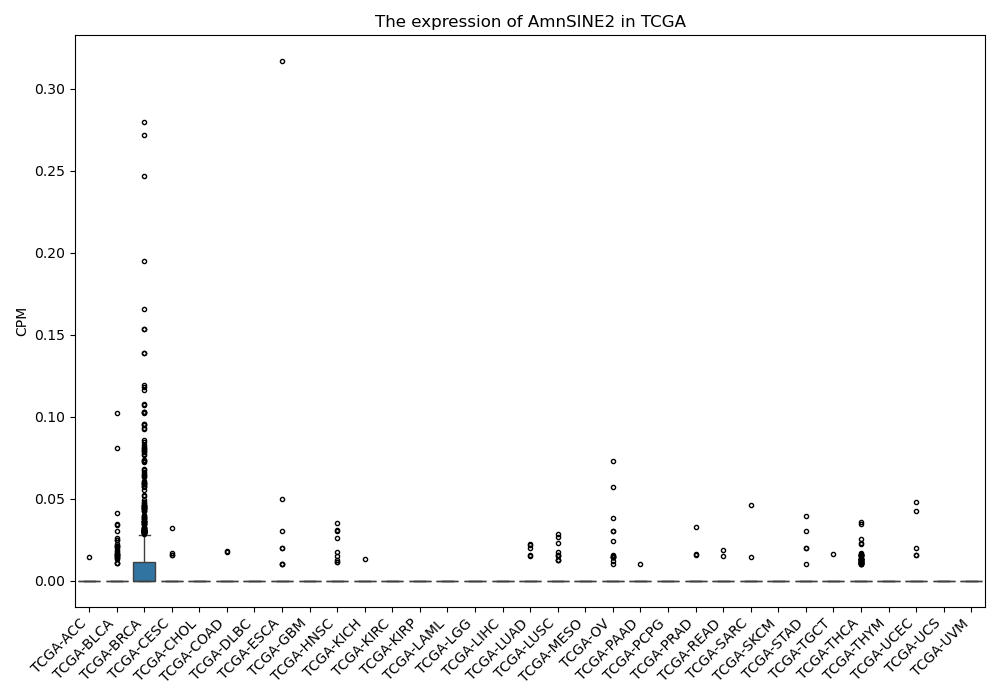
AmnSINE2
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000068 |
|---|---|
| TE superfamily | Unknown_LINE-dependent |
| TE class | SINE |
| Species | Amniota |
| Length | 358 |
| Kimura value | 34.72 |
| Tau index | 0.0000 |
| Description | Aminiota SINE2 |
| Comment | The copy number ranges from ~100 in the human genome to ~178 in the chicken genome. 45% of human copies are in highly conserved regions. Shows distant similarity to Leu tRNA. Tentatively classified as SINE element. Inverted from UCON3 original. Reference coords 1-78:tRNA-Leu- TTG, 137-264:Deu core, remainder must match an as yet unknown LINE. |
| Sequence |
GCCGGAGGGGATGGCTTAGTGGTCTAAGCATCAGGTTTGAAATACCTAGACTCCCTGGAACCACAGGTTCAAATCCCAGCAGGGTCGACTCAGCCCTTCATCCTTCCGAGGTAGATAAATTGAGTNCCACGCAGTTTACTGTGTGNGGGTCTTTCGGATGAGACCTTAAAAACCAAAGCCCTGTCTGCTCTGCGTGGACATTAAAGATCCCGTGGCGCTTTTCGTAAGAGTAGGGGTTTGCCCCGGTGCCCTTGGCCGAATTTCTGGAACCCGGCTGAAANCCCGGGGAAACCGAGAGCCTGGCTTATCCCTATAGGGTATTAGNCACCCTATAAAAGTTAAATTTATTTATTTATTT
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| AmnSINE2 | CEP3 | 104 | 110 | - | 13.09 | CTCGGAA |
| AmnSINE2 | Crg-1 | 345 | 355 | - | 13.07 | TAAATAAATAA |
| AmnSINE2 | ZBTB12 | 264 | 270 | + | 12.79 | CTGGAAC |
| AmnSINE2 | ZBTB12 | 55 | 61 | + | 12.79 | CTGGAAC |
| AmnSINE2 | CG12605 | 61 | 69 | - | 12.79 | AACCTGTGG |
| AmnSINE2 | ZBTB26 | 262 | 269 | - | 12.76 | TTCCAGAA |
| AmnSINE2 | brk | 212 | 219 | + | 12.70 | GTGGCGCT |
| AmnSINE2 | Six4 | 31 | 37 | - | 12.63 | AACCTGA |
| AmnSINE2 | TFAP2A | 1 | 11 | - | 12.62 | TCCCCTCCGGC |
| AmnSINE2 | NHP10 | 282 | 289 | + | 12.60 | CCCGGGGA |
TFBS enrichment in GRCh38
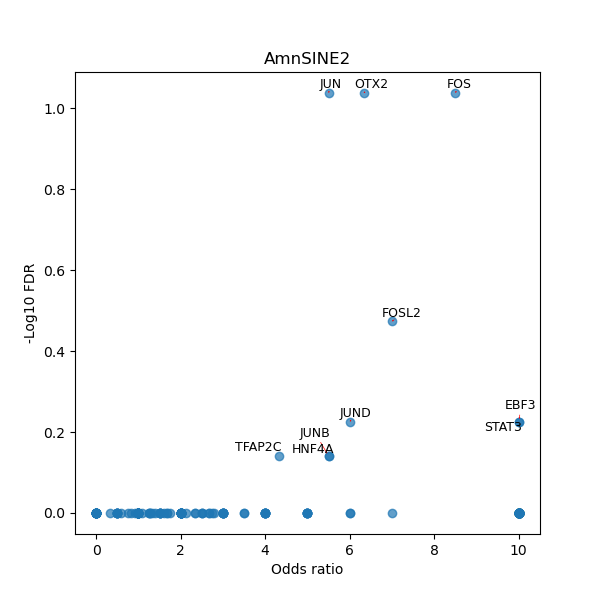
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.