TE_91
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| TE family | MIRb |
|---|---|
| TE superfamily | L2-end |
| TE class | SINE |
| Gene | - |
| Class | Intergenic |
| Coding | Non-coding |
| Tumor-specific score | 8.82 |
| Counts of samples | TCGA_tumor:1001; TCGA_normal:25; GTEx:102 |
| TSS | chr1(-):9417539 |
Transcript isoforms
| Transcript_ID | Gene | Class | Coding | ORF | Position |
|---|---|---|---|---|---|
| teRNA_91.1 | - | Intergenic | Non-coding | Non-coding | chr1(-):9411039,9411059,9414350,9414586,9417482,9417540 |
| teRNA_91.2 | - | Intergenic | Non-coding | Non-coding | chr1(-):9405006,9405026,9406968,9407089,9414350,9414586,9417482,9417540 |
| teRNA_91.3 | - | Intergenic | Non-coding | Non-coding | chr1(-):9373262,9373282,9414350,9414586,9417482,9417540 |
| teRNA_91.4 | - | Intergenic | Non-coding | Non-coding | chr1(-):9372368,9372388,9414350,9414586,9417482,9417540 |
| teRNA_91.5 | - | Intergenic | Non-coding | Non-coding | chr1(-):9405006,9405026,9414350,9414586,9417482,9417540 |
| teRNA_91.6 | - | Intergenic | Non-coding | Non-coding | chr1(-):9372368,9372388,9373141,9373282,9414350,9414586,9417482,9417540 |
| teRNA_91.7 | - | Intergenic | Non-coding | Non-coding | chr1(-):9382866,9382886,9414350,9414586,9417482,9417540 |
| teRNA_91.8 | - | Intergenic | Non-coding | Non-coding | chr1(-):9372368,9372388,9373141,9373377,9414350,9414586,9417482,9417540 |
| teRNA_91.9 | - | Intergenic | Non-coding | Non-coding | chr1(-):9409611,9409631,9414350,9414586,9417482,9417540 |
| teRNA_91.10 | - | Intergenic | Non-coding | Non-coding | chr1(-):9400276,9400296,9414350,9414586,9417482,9417540 |
1
GTEx
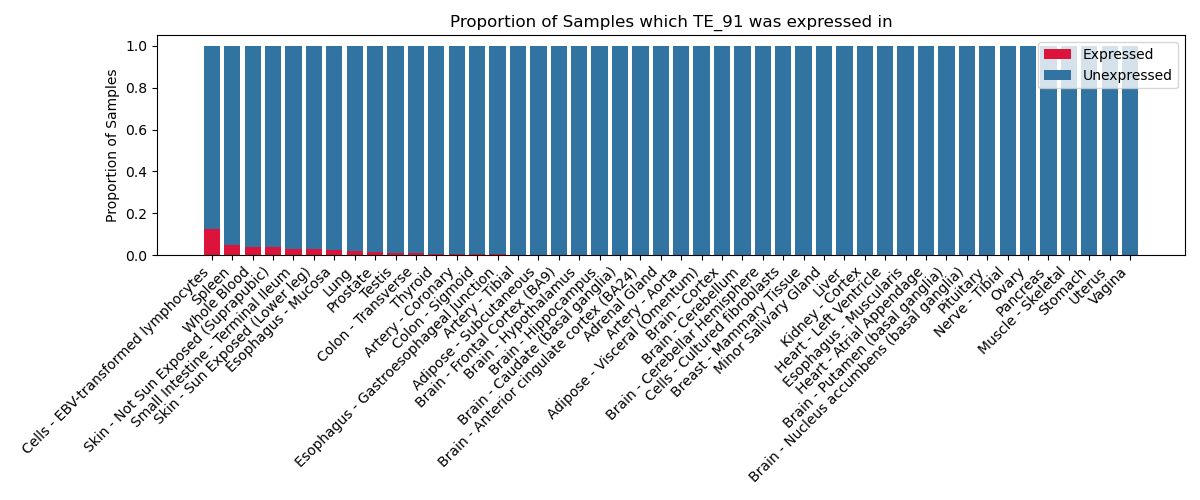
The proportion of samples in which the TE-initiated transcript was expressed across 46 body sites in the Genotype-Tissue Expression (GTEx) project.
TCGA
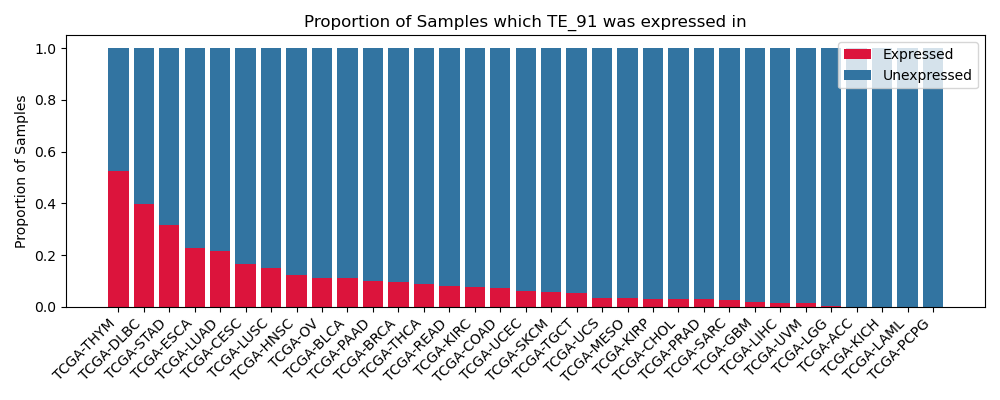
The proportion of samples in which the TE-initiated transcript was expressed across 33 cancer types from The Cancer Genome Atlas (TCGA).
GTEx
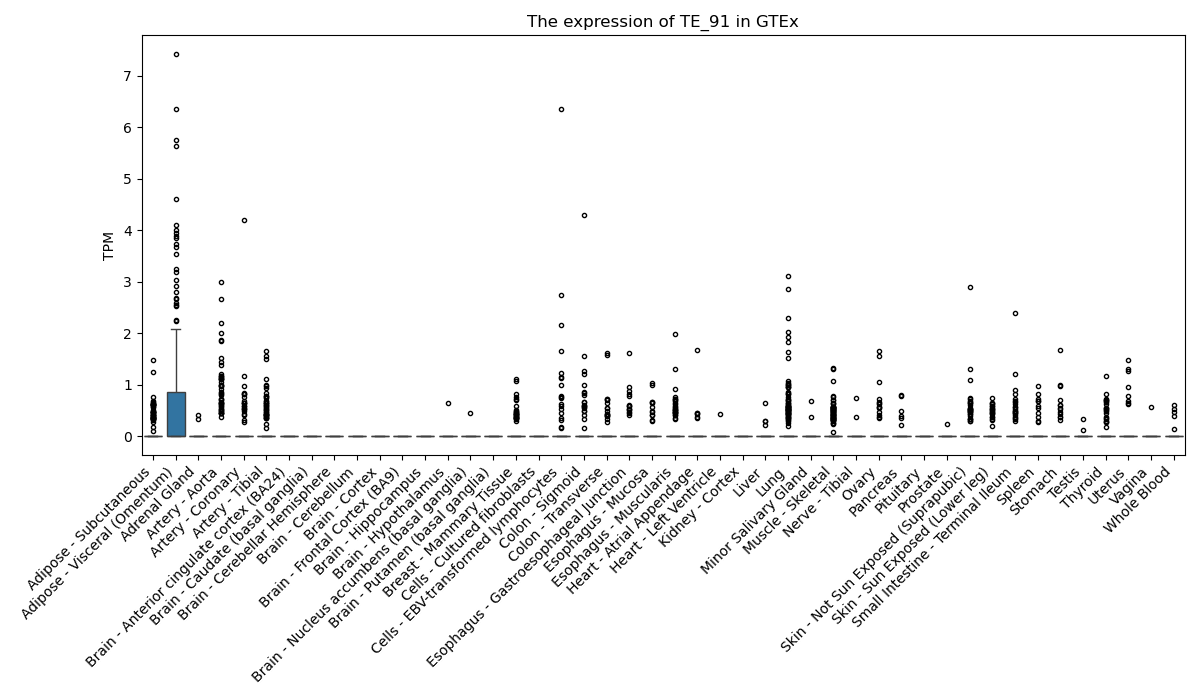
The expression of TE-initiated transcript across 46 body sites from The Genotype-Tissue Expression (GTEx) project.