TE_50
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| TE family | L1MC5a_3end |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Gene | NPHP4 |
| Class | Chimeric |
| Coding | Non-coding, Other |
| Tumor-specific score | 0.95 |
| Counts of samples | TCGA_tumor:695; TCGA_normal:28; GTEx:795 |
| TSS | chr1(-):5896813 |
Transcript isoforms
| Transcript_ID | Gene | Class | Coding | ORF | Position |
|---|---|---|---|---|---|
| teRNA_50.1 | NPHP4 | Chimeric | Other | MVGPGFLKPGERRCFARYLAVQTLQIDVWDGDSLLLIGSAAVQMKHLLRQGRPAVQASHELEVVATEYEQDNMVVSGDMLGFGRVKPIGVHSVVKGRLHLTLANVGHPCEQKVRGCSTLPPSRSRVISNDGASRFSGGSLLTTGSSRRKHVVQAQKLADVDSELAAMLLTHARQGKGPQDVSRESDATRRRKLERMRSVRLQEAGGDLGRRGTSVLAQQSVRTQHLRDLQVIAAYRERTKAESIASLLSLAITTEHTLHATLGVAEFFEFVLKNPHNTQHTVTVEIDNPELSVIVDSQEWRDFKGAAGLHTPVEEDMFHLRGSLAPQLYLRPHETAHVPFKFQSFSAGQLAMVQASPGLSNEKGMDAVSPWKSSAVPTKHAKVLFRASGGKPIAVLCLTVELQPHVVDQVFRFYHPELSFLKKAIRLPPWHTFPGAPVGMLGEDPPVHVRCSDPNVICETQNVGPGEPRDIFLKVASGPSPEIKDFFVIIYSDRWLATPTQTWQVYLHSLQRVDVSCVAGQLTRLSLVLRGTQTVRKVRAFTSHPQELKTDPKGVFVLPPRGVQDLHVGVRPLRAGSRFVHLNLVDVDCHQLVASWLVCLCCRQPLISKAFEIMLAAGEGKGVNKRITYTNPYPSRRTFHLHSDHPELLRFREDSFQVGGGETY | chr1(-):5863385,5863405,5863890,5864033,5864338,5864517,5865102,5865273,5866373,5866458,5867030,5867115,5867740,5867896,5873252,5873335,5874471,5874657,5874874,5875100,5877093,5877298,5880114,5880239,5887286,5887466,5890868,5891028,5896655,5896814 |
| teRNA_50.2 | NPHP4 | Chimeric | Other | MFHLRGSLAPQLYLRPHETAHVPFKFQSFSAGQLAMVQASPGLSNEKGMDAVSPWKSSAVPTKHAKVLFRASGGKPIAVLCLTVELQPHVVDQVFRFYHPELSFLKKAIRLPPWHTFPGAPVGMLGEDPPVHVRCSDPNVICETQNVGPGEPRDIFLKVASGPSPEIKDFFVIIYSDRWLATPTQTWQVYLHSLQRVDVSCVAGQLTRLSLVLRGTQTVRKVRAFTSHPQELKTDPKGVFVLPPRGVQDLHVGVRPLRAGSRFVHLNLVDVDCHQLVASWLVCLCCRQPLISKAFEIMLAAGEGKGVNKRITYTNPYPSRRTFHLHSDHPELLRFREDSFQVGGGETY | chr1(-):5863385,5863405,5863890,5864033,5864338,5864517,5865102,5865273,5866373,5866458,5867030,5867115,5867740,5867896,5873252,5873335,5874471,5874657,5874874,5875100,5875954,5876078,5876386,5876523,5877093,5877298,5879346,5879632,5880114,5880239,5887286,5887466,5890868,5891028,5896655,5896814 |
| teRNA_50.3 | NPHP4 | Chimeric | Non-coding | Non-coding | chr1(-):5879612,5879632,5880114,5880239,5887286,5887466,5890868,5891028,5896655,5896814 |
| teRNA_50.4 | NPHP4 | Chimeric | Non-coding | Non-coding | chr1(-):5886952,5886972,5887286,5887466,5888460,5888610,5890868,5891028,5896655,5896814 |
| teRNA_50.5 | NPHP4 | Chimeric | Other | MVVSGDMLGFGRVKPIGVHSVVKGRLHLTLANVGHPCEQKVRGCSTLPPSRSRVISNDGASRFSGGSLLTTGSSRRKHVVQAQKLADVDSELAAMLLTHARQGKGPQDVSRESDATRRRKLERMRSVRLQEAGGDLGRRGTSVLAQQSVRTQHLRDLQVIAAYRERTKAESIASLLSLAITTEHTLHATLGVAEFFEFVLKNPHNTQHTVTVEIDNPELSVIVDSQEWRDFKGAAGLHTPVEEDMFHLRGSLAPQLYLRPHETAHVPFKFQSFSAGQLAMVQASPGLSNEKGMDAVSPWKSSAVPTKHAKVLFRASGGKPIAVLCLTVELQPHVVDQVFRFYHPELSFLKKAIRLPPWHTFPGAPVGMLGEDPPVHVRCSDPNVICETQNVGPGEPRDIFLKVASGPSPEIKDFFVIIYSDRWLATPTQTWQVYLHSLQRVDVSCVAGQLTRLSLVLRGTQTVRKVRAFTSHPQELKTDPKGVFVLPPRGVQDLHVGVRPLRAGSRFVHLNLVDVDCHQLVASWLVCLCCRQPLISKAFEIMLAAGEGKGVNKRITYTNPYPSRRTFHLHSDHPELLRFREDSFQVGGGETY | chr1(-):5863385,5863405,5863890,5864033,5864338,5864517,5865102,5865273,5866373,5866458,5867030,5867115,5867740,5867896,5873252,5873335,5874471,5874657,5874874,5875100,5877093,5877298,5880114,5880239,5887286,5887466,5896655,5896814 |
| teRNA_50.6 | NPHP4 | Chimeric | Other | MFHLRGSLAPQLYLRPHETAHVPFKFQSFSAGQLAMVQASPGLSNEKGMDAVSPWKSSAVPTKHAKVLFRASGGKPIAVLCLTVELQPHVVDQVFRFYHPELSFLKKAIRLPPWHTFPGAPVGMLGEDPPVHVRCSDPNVICETQNVGPGEPRDIFLKVASGPSPEIKDFFVIIYSDRWLATPTQTWQVYLHSLQRVDVSCVAGQLTRLSLVLRGTQTVRKVRAFTSHPQELKTDPKGVFVLPPRGVQDLHVGVRPLRAGSRFVHLNLVDVDCHQLVASWLVCLCCRQPLISKAFEIMLAAGEGKGVNKRITYTNPYPSRRTFHLHSDHPELLRFREDSFQVGGGETY | chr1(-):5863385,5863405,5863890,5864033,5864338,5864517,5865102,5865273,5866373,5866458,5867030,5867115,5867740,5867896,5873252,5873335,5874471,5874657,5874874,5875100,5875954,5876078,5876386,5876523,5877093,5877298,5879346,5879632,5880114,5880239,5887286,5887466,5896655,5896814 |
| teRNA_50.7 | NPHP4 | Chimeric | Non-coding | Non-coding | chr1(-):5879612,5879632,5880114,5880239,5887286,5887466,5896655,5896814 |
| teRNA_50.8 | NPHP4 | Chimeric | Non-coding | Non-coding | chr1(-):5886952,5886972,5887286,5887466,5896655,5896814 |
| teRNA_50.9 | NPHP4 | Chimeric | Other | MLLTHARQGKGPQDVSRESDATRRRKLERMRSVRLQEAGGDLGRRGTSVLAQQSVRTQHLRDLQVIAAYRERTKAESIASLLSLAITTEHTLHATLGVAEFFEFVLKNPHNTQHTVTVEIDNPELSVIVDSQEWRDFKGAAGLHTPVEEDMFHLRGSLAPQLYLRPHETAHVPFKFQSFSAGQLAMVQASPGLSNEKGMDAVSPWKSSAVPTKHAKVLFRASGGKPIAVLCLTVELQPHVVDQVFRFYHPELSFLKKAIRLPPWHTFPGAPVGMLGEDPPVHVRCSDPNVICETQNVGPGEPRDIFLKVASGPSPEIKDFFVIIYSDRWLATPTQTWQVYLHSLQRVDVSCVAGQLTRLSLVLRGTQTVRKVRAFTSHPQELKTDPKGVFVLPPRGVQDLHVGVRPLRAGSRFVHLNLVDVDCHQLVASWLVCLCCRQPLISKAFEIMLAAGEGKGVNKRITYTNPYPSRRTFHLHSDHPELLRFREDSFQVGGGETY | chr1(-):5863385,5863405,5863890,5864033,5864338,5864517,5865102,5865273,5866373,5866458,5867030,5867115,5867740,5867896,5873252,5873335,5874471,5874657,5874874,5875100,5877093,5877298,5880114,5880239,5896655,5896814 |
| teRNA_50.10 | NPHP4 | Chimeric | Other | MFHLRGSLAPQLYLRPHETAHVPFKFQSFSAGQLAMVQASPGLSNEKGMDAVSPWKSSAVPTKHAKVLFRASGGKPIAVLCLTVELQPHVVDQVFRFYHPELSFLKKAIRLPPWHTFPGAPVGMLGEDPPVHVRCSDPNVICETQNVGPGEPRDIFLKVASGPSPEIKDFFVIIYSDRWLATPTQTWQVYLHSLQRVDVSCVAGQLTRLSLVLRGTQTVRKVRAFTSHPQELKTDPKGVFVLPPRGVQDLHVGVRPLRAGSRFVHLNLVDVDCHQLVASWLVCLCCRQPLISKAFEIMLAAGEGKGVNKRITYTNPYPSRRTFHLHSDHPELLRFREDSFQVGGGETY | chr1(-):5863385,5863405,5863890,5864033,5864338,5864517,5865102,5865273,5866373,5866458,5867030,5867115,5867740,5867896,5873252,5873335,5874471,5874657,5874874,5875100,5875954,5876078,5876386,5876523,5877093,5877298,5879346,5879632,5880114,5880239,5896655,5896814 |
1
2
GTEx
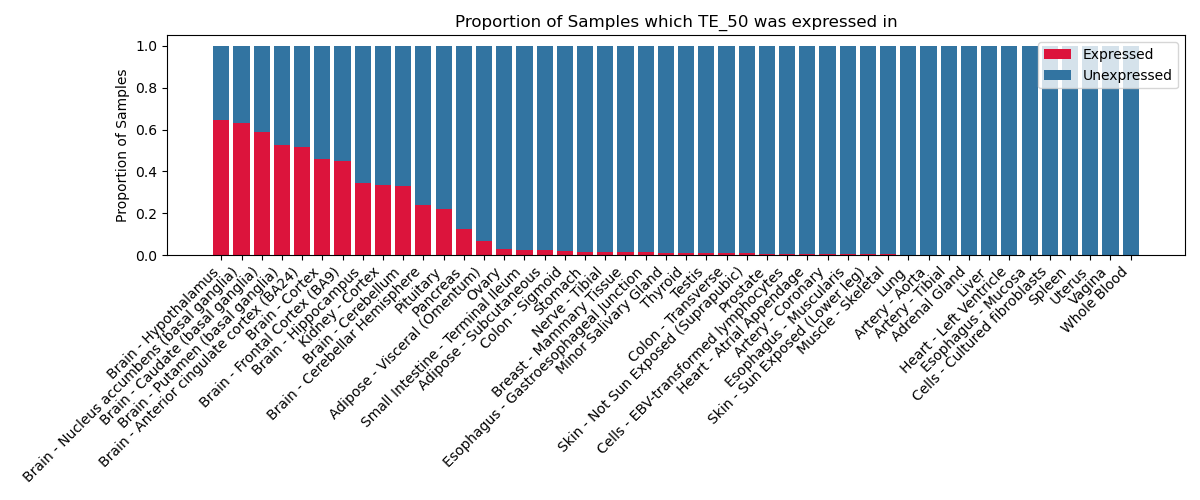
The proportion of samples in which the TE-initiated transcript was expressed across 46 body sites in the Genotype-Tissue Expression (GTEx) project.
TCGA
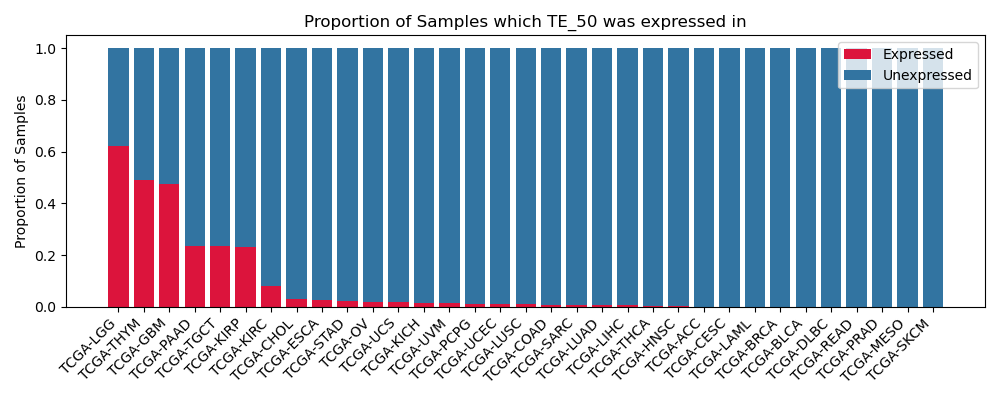
The proportion of samples in which the TE-initiated transcript was expressed across 33 cancer types from The Cancer Genome Atlas (TCGA).
GTEx
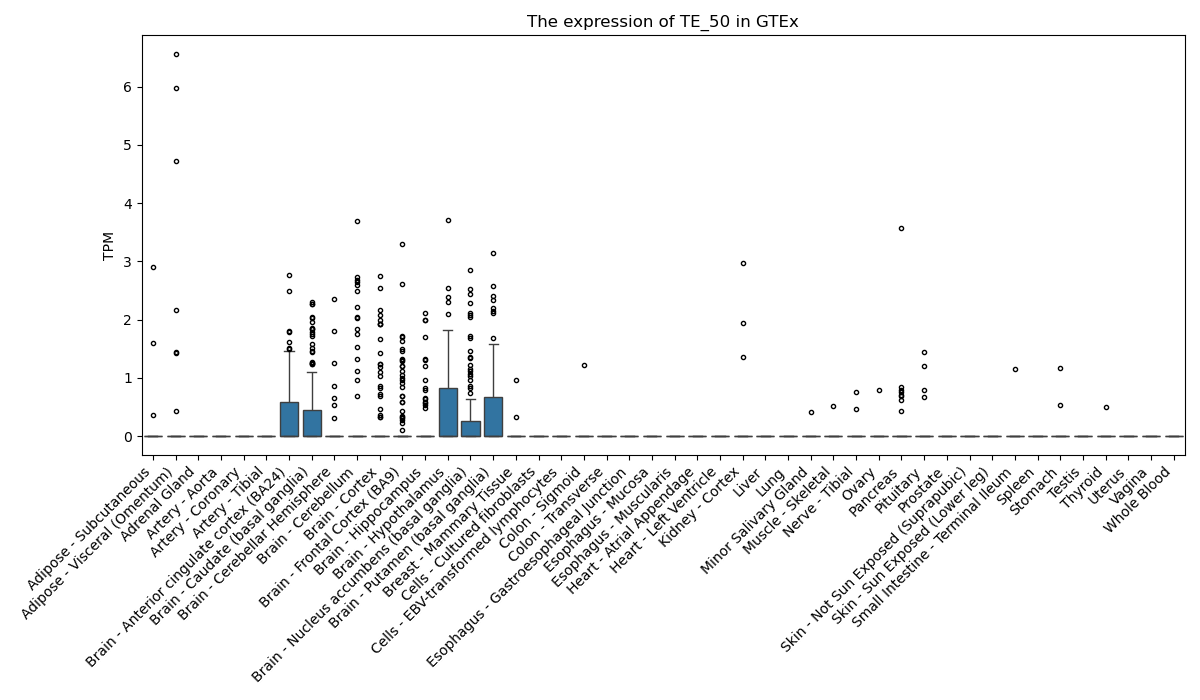
The expression of TE-initiated transcript across 46 body sites from The Genotype-Tissue Expression (GTEx) project.