TE_159
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| TE family | L2a_3end |
|---|---|
| TE superfamily | L2 |
| TE class | LINE |
| Gene | CROCCP3 |
| Class | Chimeric |
| Coding | Non-coding, Other |
| Tumor-specific score | 0.31 |
| Counts of samples | TCGA_tumor:189; TCGA_normal:25; GTEx:674 |
| TSS | chr1(-):16497656 |
Transcript isoforms
| Transcript_ID | Gene | Class | Coding | ORF | Position |
|---|---|---|---|---|---|
| teRNA_159.1 | CROCCP3 | Chimeric | Other | MRGRYEASQDLLGTLRKQLSDSESERRALEEQLQRLRDKTDSTMQAHEDAQREVQRLRSAKELLRREKSNLAHSLQVAQQQAKELRQERKKLQAAQEELRRQRYWLGEEQEDAVQDGVRVRRELEHSHRQLEQLEGKRSVLAKELVEVREALSRATLQRDMLQAEKAEVAEALTKAEGRGGLPAGLPVQAERPQREPCSGQVGSEPPCHPGHSWRKKSAPCRAGSGRRSRRPQWHGKSRSGWRRCGWNRRWRGRAWRAPYERRSRPRRQWSSSSPRCIMSAAGCRSS* | chr1(-):16470107,16470127,16475825,16476062,16476399,16476504,16476930,16477074,16478255,16478437,16483287,16483486,16484233,16484464,16484823,16484906,16485569,16485665,16486426,16486661,16490618,16490783,16490865,16490926,16491010,16492179,16497524,16497657 |
| teRNA_159.2 | CROCCP3 | Chimeric | Other | MRGRYEASQDLLGTLRKQLSDSESERRALEEQLQRLRDKTDSTMQAHEDAQREVQRLRSAKELLRREKSNLAHSLQVAQQQAKELRQERKKLQAAQEELRRQRYWLGEEQEDAVQDGVRVRRELEHSHRQLEQLEGKRSVLAKELVEVREALSRATLQRDMLQAEKAEVAEALTKAEGRGGLPAGLPVQAERPQREPCSGQVGSEPPCHPGHSWRKKSAPCRAGSGRRSRRPQWHGKSRSGWRRCGWNRRWRGRAWRAPYERRSRPRRQWSSSSPRCIMSAAGCRSS* | chr1(-):16468987,16469007,16475825,16476062,16476399,16476504,16476930,16477074,16478255,16478437,16483287,16483486,16484233,16484464,16485569,16485665,16489685,16489791,16490618,16490783,16490865,16490926,16491010,16492179,16497524,16497657 |
| teRNA_159.3 | CROCCP3 | Chimeric | Non-coding | Non-coding | chr1(-):16490763,16490783,16490865,16490926,16491010,16492179,16497524,16497657 |
| teRNA_159.4 | CROCCP3 | Chimeric | Other | MRGRYEASQDLLGTLRKQLSDSESERRALEEQLQRLRDKTDSTMQAHEDAQREVQRLRSAKELLRREKSNLAHSLQVAQQQAKELRQERKKLQAAQEELRRQRYWLGEEQEDAVQDGVRVRRELEHSHRQLEQLEGKRSVLAKELVEVREALSRATLQRDMLQAEKAEVAEALTKAEGRGGLPAGLPVQAERPQREPCSGQVGSEPPCHPGHSWRKKSAPCRAGSGRRSRRPQWHGKSRSGWRRCGWNRRWRGRAWRAPYERRSRPRRQWSSSSPRCIMSAAGCRSS* | chr1(-):16474469,16474489,16475825,16476062,16476399,16476504,16476930,16477074,16478255,16478437,16483287,16483486,16484233,16484464,16484823,16484906,16485569,16485665,16486426,16486661,16489685,16489791,16490618,16490783,16490865,16490926,16491010,16492179,16497524,16497657 |
| teRNA_159.5 | CROCCP3 | Chimeric | Other | MRGRYEASQDLLGTLRKQLSDSESERRALEEQLQRLRDKTDSTMQAHEDAQREVQRLRSAKELLRREKSNLAHSLQVAQQQAKELRQERKKLQAAQEELRRQRYWLGEEQEDAVQDGVRVRRELEHSHRQLEQLEGKRSVLAKELVEVREALSRATLQRDMLQAEKAEVAEALTKAEGRGGLPAGLPVQAERPQREPCSGQVGSEPPCHPGHSWRKKSAPCRAGSGRRSRRPQWHGKSRSGWRRCGWNRRWRGRAWRAPYERRSRPRRQWSSSSPRCIMSAAGCRSS* | chr1(-):16470107,16470127,16475825,16476062,16476399,16476504,16476930,16477074,16478255,16478437,16483287,16483486,16484233,16484464,16484823,16484906,16485569,16485665,16486426,16486661,16490618,16490783,16490865,16490926,16491010,16491084,16491186,16491316,16491981,16492179,16497524,16497657 |
| teRNA_159.6 | CROCCP3 | Chimeric | Other | MRGRYEASQDLLGTLRKQLSDSESERRALEEQLQRLRDKTDSTMQAHEDAQREVQRLRSAKELLRREKSNLAHSLQVAQQQAKELRQERKKLQAAQEELRRQRYWLGEEQEDAVQDGVRVRRELEHSHRQLEQLEGKRSVLAKELVEVREALSRATLQRDMLQAEKAEVAEALTKAEGRGGLPAGLPVQAERPQREPCSGQVGSEPPCHPGHSWRKKSAPCRAGSGRRSRRPQWHGKSRSGWRRCGWNRRWRGRAWRAPYERRSRPRRQWSSSSPRCIMSAAGCRSS* | chr1(-):16470107,16470127,16475825,16476062,16476399,16476504,16476930,16477074,16478255,16478437,16483287,16483486,16484233,16484464,16484823,16484906,16485569,16485665,16486426,16486661,16490618,16490783,16490865,16490926,16497524,16497657 |
| teRNA_159.7 | CROCCP3 | Chimeric | Other | MRGRYEASQDLLGTLRKQLSDSESERRALEEQLQRLRDKTDSTMQAHEDAQREVQRLRSAKELLRREKSNLAHSLQVAQQQAKELRQERKKLQAAQEELRRQRYWLGEEQEDAVQDGVRVRRELEHSHRQLEQLEGKRSVLAKELVEVREALSRATLQRDMLQAEKAEVAEALTKAEGRGGLPAGLPVQAERPQREPCSGQVGSEPPCHPGHSWRKKSAPCRAGSGRRSRRPQWHGKSRSGWRRCGWNRRWRGRAWRAPYERRSRPRRQWSSSSPRCIMSAAGCRSS* | chr1(-):16468987,16469007,16475825,16476062,16476399,16476504,16476930,16477074,16478255,16478437,16483287,16483486,16484233,16484464,16485569,16485665,16489685,16489791,16490618,16490783,16490865,16490926,16497524,16497657 |
| teRNA_159.8 | CROCCP3 | Chimeric | Other | MRGRYEASQDLLGTLRKQLSDSESERRALEEQLQRLRDKTDSTMQAHEDAQREVQRLRSAKELLRREKSNLAHSLQVAQQQAKELRQERKKLQAAQEELRRQRYWLGEEQEDAVQDGVRVRRELEHSHRQLEQLEGKRSVLAKELVEVREALSRATLQRDMLQAEKAEVAEALTKAEGRGGLPAGLPVQAERPQREPCSGQVGSEPPCHPGHSWRKKSAPCRAGSGRRSRRPQWHGKSRSGWRRCGWNRRWRGRAWRAPYERRSRPRRQWSSSSPRCIMSAAGCRSS* | chr1(-):16474054,16474074,16475825,16476062,16476399,16476504,16476930,16477074,16478255,16478437,16483287,16483486,16484233,16484464,16484823,16484906,16485569,16485665,16486426,16486661,16490618,16490783,16490865,16490926,16497524,16497657 |
| teRNA_159.9 | CROCCP3 | Chimeric | Non-coding | Non-coding | chr1(-):16490763,16490783,16490865,16490926,16497524,16497657 |
| teRNA_159.10 | CROCCP3 | Chimeric | Other | MRGRYEASQDLLGTLRKQLSDSESERRALEEQLQRLRDKTDSTMQAHEDAQREVQRLRSAKELLRREKSNLAHSLQVAQQQAKELRQERKKLQAAQEELRRQRYWLGEEQEDAVQDGVRVRRELEHSHRQLEQLEGKRSVLAKELVEVREALSRATLQRDMLQAEKAEVAEALTKAEGRGGLPAGLPVQAERPQREPCSGQVGSEPPCHPGHSWRKKSAPCRAGSGRRSRRPQWHGKSRSGWRRCGWNRRWRGRAWRAPYERRSRPRRQWSSSSPRCIMSAAGCRSS* | chr1(-):16474469,16474489,16475825,16476062,16476399,16476504,16476930,16477074,16478255,16478437,16483287,16483486,16484233,16484464,16484823,16484906,16485569,16485665,16486426,16486661,16489685,16489791,16490618,16490783,16490865,16490926,16497524,16497657 |
GTEx
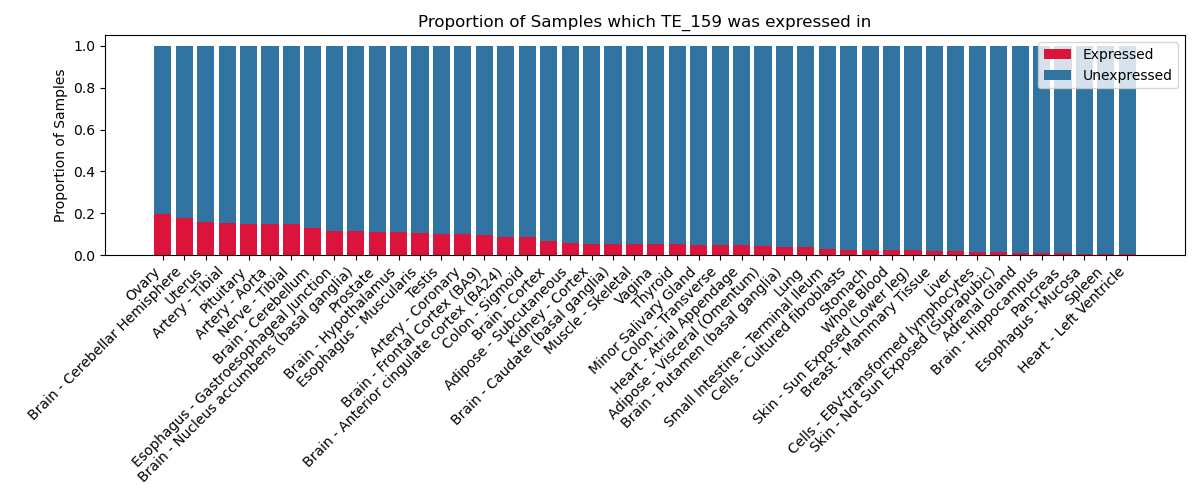
The proportion of samples in which the TE-initiated transcript was expressed across 46 body sites in the Genotype-Tissue Expression (GTEx) project.
TCGA
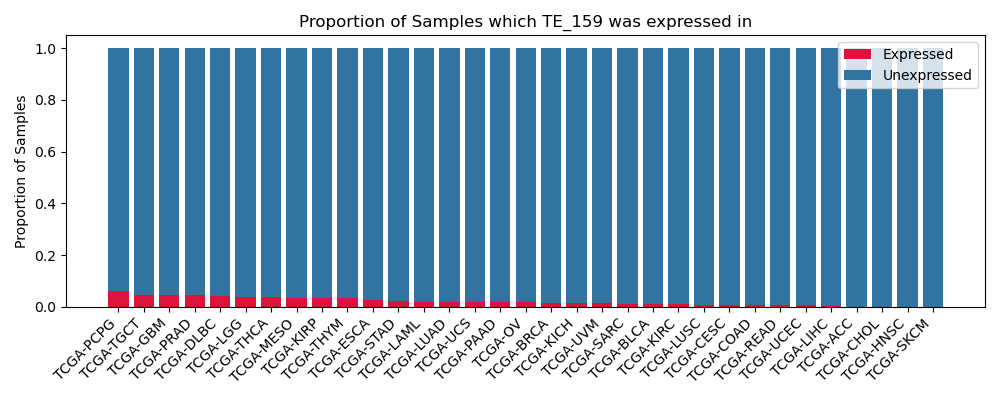
The proportion of samples in which the TE-initiated transcript was expressed across 33 cancer types from The Cancer Genome Atlas (TCGA).
GTEx
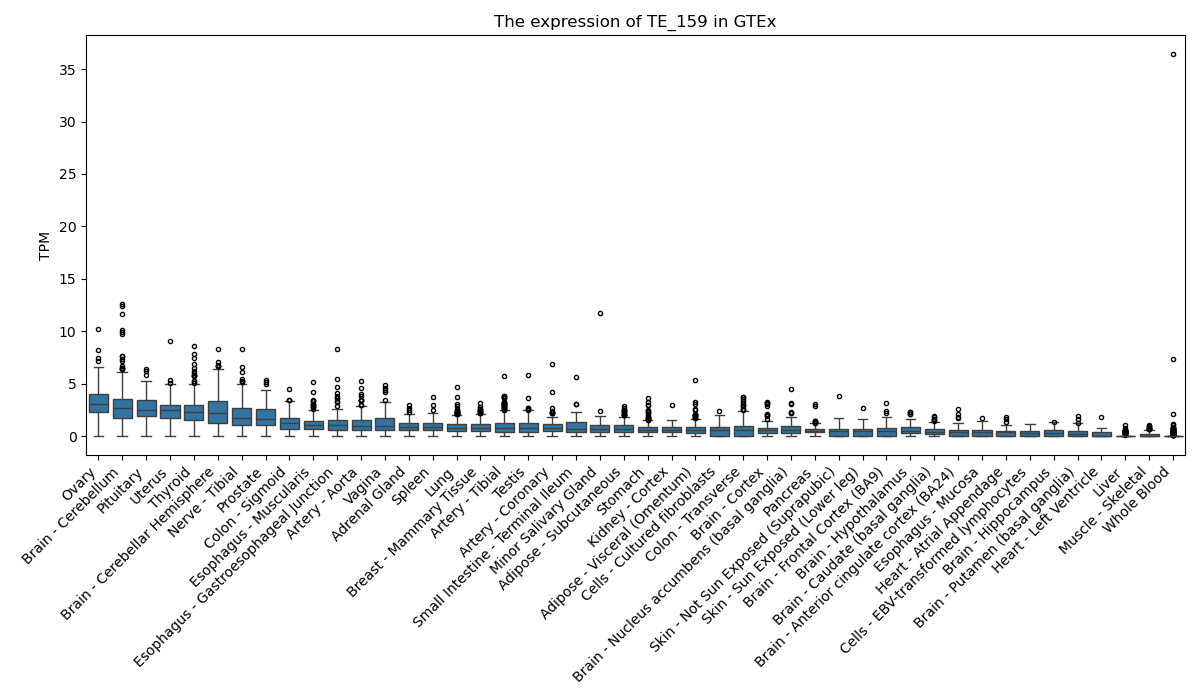
The expression of TE-initiated transcript across 46 body sites from The Genotype-Tissue Expression (GTEx) project.