hAT-4b_Ther
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001152 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Theria_mammals |
| Length | 1263 |
| Kimura value | 33.43 |
| Tau index | 0.0000 |
| Description | hAT-Tip100 DNA transposon, hAT-4b_Ther subfamily |
| Comment | Present in placentals and marsupials, absent from monotremes. Nearly identical to hAT-4_Crp (which was active in an ancestor of all crocodiles) except for numerous small and one 2160 bp deletion (at pos 997-998). |
| Sequence |
AGGGCCNAAGCGTGGNGNAGTGGGAAGGGTACGCTTTCTAGGNCCCGAAGATNCAGGGGGNCCCAGCCCAGGCTGCAACCCTATTACCACGAGGAAAAAAACTAAGGCTAAATTNTAGGTCATGCTTGAGGATATTTATGGAAAGTTATGTNCTACCTATATGGGATCTGTGAGCCTGGAGCCAATCAGAAGCAGAACAAGCAACATTCTGAACAGTGAGTTGCTGCTTTCTGTAGAACAGAGAGAGTGTGCTTCTTGTTGTGCAGCTAAATCTAGGACTTGTAGTTTGAGAGACAGANAGAGGGATTTATGCTTCCCTATGTGACATGCTCTCTCAACTACAAGTCCCAGAGTCCCTTGTGCATGGAGAGGGGAGGGACAAAGCACCACATAACCAGGGACATTCCCAGAAAGGACAGCGGAAGCCACAGCGCCGCTTCTGCTGGAGGATTTACAGGCCCAAAANCGAGTAATGAGTTAGGCCTGGCGGTCTACACGTTGCACAGAGAGAAATNAAAATCAAAACAAAANAGGTAGATTAGTGCCTGTGGGAAGGAAAGGACATCTCTTTCTCTCTCTCTCACACGCGCACCATGTCTGGCTATAAGTACAAATCAGGCAGCCAGAAACATAAGGAGGTAAAACAAAGGGAACTAAAAGAAGCAAAAGGACACCAATCAATCCTCCAGTTTTTCAAATCAGACAAGGCCCTGGCAACTGGTGATGATGAAGAAAAGCTGCTGCCAGATGCATCATGTGATGACTCCAATGATCTAATTCTTTCATCATCAAAAGAGAGTCAGGTAATATCACCATTAGCAGAAGAAGGCTTAAAAAAACAGGAGAGAGNAGTAGTACTAGTACTACACTATCTGATTCATGCTTTTTCAAAGACCCAGGCTTGTGGCCTAAACTGATAACGGATTCTATCCATCAAAAAATTGTTCTATCTGGTGTCCTGACTTTTGAAGAAATGCGGAAGCTAGCAAAGTGAATTGGTCTATTTATAAAATGTTTGCAGCCATGTTGGTCCATTAGAACAACAAACAAATCAAAACATCGACTGGATAATACCGTTATTAGGACCAACAACCCAAAAGTTACAAAATAGTCAGCAATGTAGAATGTAATTTCAGAATTAAAATAGAAATTTTTCTTTCATGATTCATGAGATTATGTATGTGGGATGGGATGGTGTGTGTAGAAGGGGGCGCACCTTACAANTTTAGACTGGGCGGGCCCGAAGAAATNATGCTTGGGCCC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| hAT-4b_Ther | Trl | 572 | 580 | - | 14.81 | GAGAGAGAG |
| hAT-4b_Ther | Trl | 574 | 582 | - | 14.81 | GAGAGAGAG |
| hAT-4b_Ther | KLF15 | 1233 | 1240 | - | 14.65 | GCCCGCCC |
| hAT-4b_Ther | MYB56 | 1071 | 1080 | - | 14.64 | ATAACGGTAT |
| hAT-4b_Ther | ZBTB6 | 175 | 183 | + | 14.62 | CCTGGAGCC |
| hAT-4b_Ther | NFYA | 182 | 189 | + | 14.54 | CCAATCAG |
| hAT-4b_Ther | dsf | 960 | 967 | - | 14.52 | AAAAGTCA |
| hAT-4b_Ther | AGL27 | 1138 | 1151 | - | 14.50 | ATTTCTATTTTAAT |
| hAT-4b_Ther | SBP6 | 606 | 613 | + | 14.34 | AAGTACAA |
| hAT-4b_Ther | eor-1 | 571 | 583 | - | 14.34 | TGAGAGAGAGAGA |
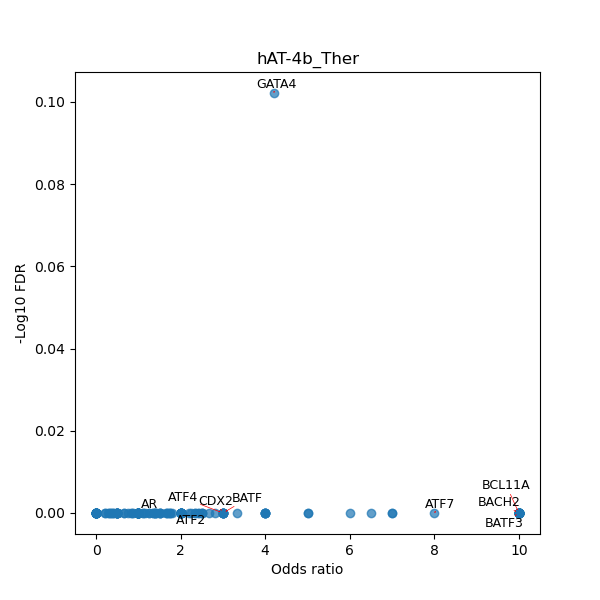
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.