hAT-4b_Ther
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001152 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Theria_mammals |
| Length | 1263 |
| Kimura value | 33.43 |
| Tau index | 0.0000 |
| Description | hAT-Tip100 DNA transposon, hAT-4b_Ther subfamily |
| Comment | Present in placentals and marsupials, absent from monotremes. Nearly identical to hAT-4_Crp (which was active in an ancestor of all crocodiles) except for numerous small and one 2160 bp deletion (at pos 997-998). |
| Sequence |
AGGGCCNAAGCGTGGNGNAGTGGGAAGGGTACGCTTTCTAGGNCCCGAAGATNCAGGGGGNCCCAGCCCAGGCTGCAACCCTATTACCACGAGGAAAAAAACTAAGGCTAAATTNTAGGTCATGCTTGAGGATATTTATGGAAAGTTATGTNCTACCTATATGGGATCTGTGAGCCTGGAGCCAATCAGAAGCAGAACAAGCAACATTCTGAACAGTGAGTTGCTGCTTTCTGTAGAACAGAGAGAGTGTGCTTCTTGTTGTGCAGCTAAATCTAGGACTTGTAGTTTGAGAGACAGANAGAGGGATTTATGCTTCCCTATGTGACATGCTCTCTCAACTACAAGTCCCAGAGTCCCTTGTGCATGGAGAGGGGAGGGACAAAGCACCACATAACCAGGGACATTCCCAGAAAGGACAGCGGAAGCCACAGCGCCGCTTCTGCTGGAGGATTTACAGGCCCAAAANCGAGTAATGAGTTAGGCCTGGCGGTCTACACGTTGCACAGAGAGAAATNAAAATCAAAACAAAANAGGTAGATTAGTGCCTGTGGGAAGGAAAGGACATCTCTTTCTCTCTCTCTCACACGCGCACCATGTCTGGCTATAAGTACAAATCAGGCAGCCAGAAACATAAGGAGGTAAAACAAAGGGAACTAAAAGAAGCAAAAGGACACCAATCAATCCTCCAGTTTTTCAAATCAGACAAGGCCCTGGCAACTGGTGATGATGAAGAAAAGCTGCTGCCAGATGCATCATGTGATGACTCCAATGATCTAATTCTTTCATCATCAAAAGAGAGTCAGGTAATATCACCATTAGCAGAAGAAGGCTTAAAAAAACAGGAGAGAGNAGTAGTACTAGTACTACACTATCTGATTCATGCTTTTTCAAAGACCCAGGCTTGTGGCCTAAACTGATAACGGATTCTATCCATCAAAAAATTGTTCTATCTGGTGTCCTGACTTTTGAAGAAATGCGGAAGCTAGCAAAGTGAATTGGTCTATTTATAAAATGTTTGCAGCCATGTTGGTCCATTAGAACAACAAACAAATCAAAACATCGACTGGATAATACCGTTATTAGGACCAACAACCCAAAAGTTACAAAATAGTCAGCAATGTAGAATGTAATTTCAGAATTAAAATAGAAATTTTTCTTTCATGATTCATGAGATTATGTATGTGGGATGGGATGGTGTGTGTAGAAGGGGGCGCACCTTACAANTTTAGACTGGGCGGGCCCGAAGAAATNATGCTTGGGCCC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| hAT-4b_Ther | BPC6 | 564 | 584 | + | 27.03 | ATCTCTTTCTCTCTCTCTCAC |
| hAT-4b_Ther | BPC6 | 566 | 586 | + | 25.63 | CTCTTTCTCTCTCTCTCACAC |
| hAT-4b_Ther | BPC6 | 562 | 582 | + | 24.53 | ACATCTCTTTCTCTCTCTCTC |
| hAT-4b_Ther | BPC1 | 563 | 586 | - | 24.11 | GTGTGAGAGAGAGAGAAAGAGATG |
| hAT-4b_Ther | Thap11 | 338 | 351 | + | 23.34 | ACTACAAGTCCCAG |
| hAT-4b_Ther | RAMOSA1 | 569 | 582 | - | 22.37 | GAGAGAGAGAGAAA |
| hAT-4b_Ther | RAMOSA1 | 571 | 584 | - | 21.41 | GTGAGAGAGAGAGA |
| hAT-4b_Ther | BPC1 | 561 | 584 | - | 18.68 | GTGAGAGAGAGAGAAAGAGATGTC |
| hAT-4b_Ther | ZNF213 | 66 | 77 | + | 18.38 | GCCCAGGCTGCA |
| hAT-4b_Ther | Thap11 | 273 | 286 | - | 18.03 | ACTACAAGTCCTAG |
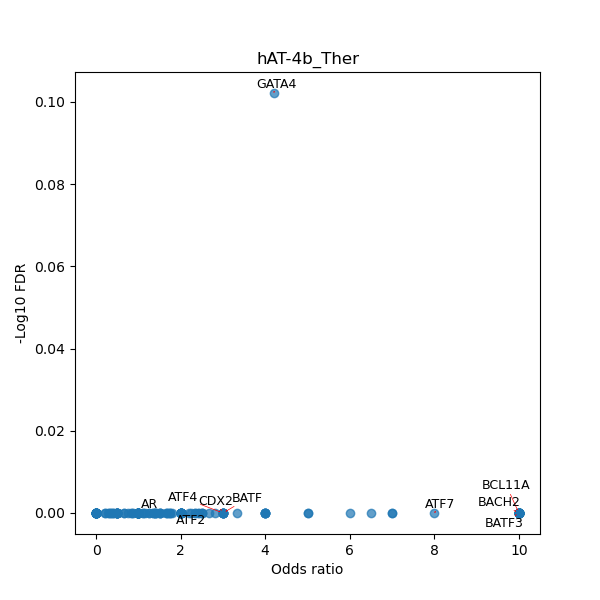
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.