hAT-4b_Ther
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001152 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Theria_mammals |
| Length | 1263 |
| Kimura value | 33.43 |
| Tau index | 0.0000 |
| Description | hAT-Tip100 DNA transposon, hAT-4b_Ther subfamily |
| Comment | Present in placentals and marsupials, absent from monotremes. Nearly identical to hAT-4_Crp (which was active in an ancestor of all crocodiles) except for numerous small and one 2160 bp deletion (at pos 997-998). |
| Sequence |
AGGGCCNAAGCGTGGNGNAGTGGGAAGGGTACGCTTTCTAGGNCCCGAAGATNCAGGGGGNCCCAGCCCAGGCTGCAACCCTATTACCACGAGGAAAAAAACTAAGGCTAAATTNTAGGTCATGCTTGAGGATATTTATGGAAAGTTATGTNCTACCTATATGGGATCTGTGAGCCTGGAGCCAATCAGAAGCAGAACAAGCAACATTCTGAACAGTGAGTTGCTGCTTTCTGTAGAACAGAGAGAGTGTGCTTCTTGTTGTGCAGCTAAATCTAGGACTTGTAGTTTGAGAGACAGANAGAGGGATTTATGCTTCCCTATGTGACATGCTCTCTCAACTACAAGTCCCAGAGTCCCTTGTGCATGGAGAGGGGAGGGACAAAGCACCACATAACCAGGGACATTCCCAGAAAGGACAGCGGAAGCCACAGCGCCGCTTCTGCTGGAGGATTTACAGGCCCAAAANCGAGTAATGAGTTAGGCCTGGCGGTCTACACGTTGCACAGAGAGAAATNAAAATCAAAACAAAANAGGTAGATTAGTGCCTGTGGGAAGGAAAGGACATCTCTTTCTCTCTCTCTCACACGCGCACCATGTCTGGCTATAAGTACAAATCAGGCAGCCAGAAACATAAGGAGGTAAAACAAAGGGAACTAAAAGAAGCAAAAGGACACCAATCAATCCTCCAGTTTTTCAAATCAGACAAGGCCCTGGCAACTGGTGATGATGAAGAAAAGCTGCTGCCAGATGCATCATGTGATGACTCCAATGATCTAATTCTTTCATCATCAAAAGAGAGTCAGGTAATATCACCATTAGCAGAAGAAGGCTTAAAAAAACAGGAGAGAGNAGTAGTACTAGTACTACACTATCTGATTCATGCTTTTTCAAAGACCCAGGCTTGTGGCCTAAACTGATAACGGATTCTATCCATCAAAAAATTGTTCTATCTGGTGTCCTGACTTTTGAAGAAATGCGGAAGCTAGCAAAGTGAATTGGTCTATTTATAAAATGTTTGCAGCCATGTTGGTCCATTAGAACAACAAACAAATCAAAACATCGACTGGATAATACCGTTATTAGGACCAACAACCCAAAAGTTACAAAATAGTCAGCAATGTAGAATGTAATTTCAGAATTAAAATAGAAATTTTTCTTTCATGATTCATGAGATTATGTATGTGGGATGGGATGGTGTGTGTAGAAGGGGGCGCACCTTACAANTTTAGACTGGGCGGGCCCGAAGAAATNATGCTTGGGCCC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| hAT-4b_Ther | Clamp | 569 | 582 | - | 15.44 | GAGAGAGAGAGAAA |
| hAT-4b_Ther | GRF4 | 737 | 744 | - | 15.23 | GCAGCAGC |
| hAT-4b_Ther | STAT3 | 404 | 412 | - | 15.20 | TTCTGGGAA |
| hAT-4b_Ther | ASR1 | 456 | 464 | + | 15.13 | AGGCCCAAA |
| hAT-4b_Ther | STAT1 | 404 | 412 | - | 15.13 | TTCTGGGAA |
| hAT-4b_Ther | YPR015C | 447 | 457 | - | 15.12 | CTGTAAATCCT |
| hAT-4b_Ther | Stat5a::Stat5b | 404 | 412 | + | 15.12 | TTCCCAGAA |
| hAT-4b_Ther | Spz1 | 72 | 82 | - | 15.12 | AGGGTTGCAGC |
| hAT-4b_Ther | BPC1 | 565 | 588 | - | 15.11 | GCGTGTGAGAGAGAGAGAAAGAGA |
| hAT-4b_Ther | HAP3 | 180 | 188 | - | 15.11 | TGATTGGCT |
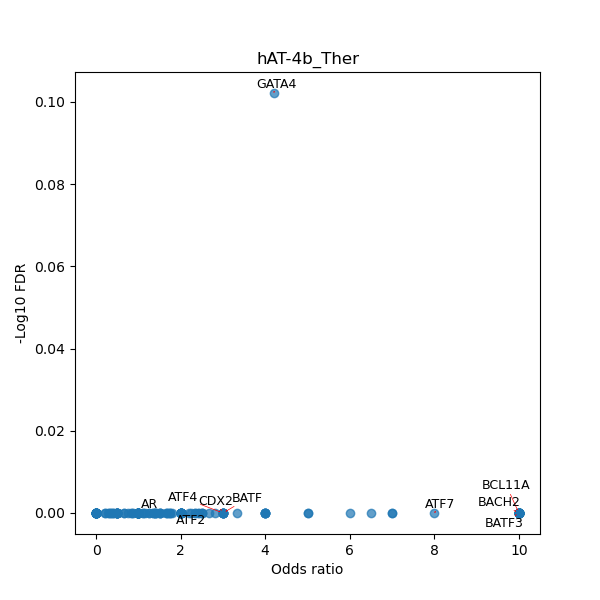
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.