hAT-4b_Ther
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001152 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Theria_mammals |
| Length | 1263 |
| Kimura value | 33.43 |
| Tau index | 0.0000 |
| Description | hAT-Tip100 DNA transposon, hAT-4b_Ther subfamily |
| Comment | Present in placentals and marsupials, absent from monotremes. Nearly identical to hAT-4_Crp (which was active in an ancestor of all crocodiles) except for numerous small and one 2160 bp deletion (at pos 997-998). |
| Sequence |
AGGGCCNAAGCGTGGNGNAGTGGGAAGGGTACGCTTTCTAGGNCCCGAAGATNCAGGGGGNCCCAGCCCAGGCTGCAACCCTATTACCACGAGGAAAAAAACTAAGGCTAAATTNTAGGTCATGCTTGAGGATATTTATGGAAAGTTATGTNCTACCTATATGGGATCTGTGAGCCTGGAGCCAATCAGAAGCAGAACAAGCAACATTCTGAACAGTGAGTTGCTGCTTTCTGTAGAACAGAGAGAGTGTGCTTCTTGTTGTGCAGCTAAATCTAGGACTTGTAGTTTGAGAGACAGANAGAGGGATTTATGCTTCCCTATGTGACATGCTCTCTCAACTACAAGTCCCAGAGTCCCTTGTGCATGGAGAGGGGAGGGACAAAGCACCACATAACCAGGGACATTCCCAGAAAGGACAGCGGAAGCCACAGCGCCGCTTCTGCTGGAGGATTTACAGGCCCAAAANCGAGTAATGAGTTAGGCCTGGCGGTCTACACGTTGCACAGAGAGAAATNAAAATCAAAACAAAANAGGTAGATTAGTGCCTGTGGGAAGGAAAGGACATCTCTTTCTCTCTCTCTCACACGCGCACCATGTCTGGCTATAAGTACAAATCAGGCAGCCAGAAACATAAGGAGGTAAAACAAAGGGAACTAAAAGAAGCAAAAGGACACCAATCAATCCTCCAGTTTTTCAAATCAGACAAGGCCCTGGCAACTGGTGATGATGAAGAAAAGCTGCTGCCAGATGCATCATGTGATGACTCCAATGATCTAATTCTTTCATCATCAAAAGAGAGTCAGGTAATATCACCATTAGCAGAAGAAGGCTTAAAAAAACAGGAGAGAGNAGTAGTACTAGTACTACACTATCTGATTCATGCTTTTTCAAAGACCCAGGCTTGTGGCCTAAACTGATAACGGATTCTATCCATCAAAAAATTGTTCTATCTGGTGTCCTGACTTTTGAAGAAATGCGGAAGCTAGCAAAGTGAATTGGTCTATTTATAAAATGTTTGCAGCCATGTTGGTCCATTAGAACAACAAACAAATCAAAACATCGACTGGATAATACCGTTATTAGGACCAACAACCCAAAAGTTACAAAATAGTCAGCAATGTAGAATGTAATTTCAGAATTAAAATAGAAATTTTTCTTTCATGATTCATGAGATTATGTATGTGGGATGGGATGGTGTGTGTAGAAGGGGGCGCACCTTACAANTTTAGACTGGGCGGGCCCGAAGAAATNATGCTTGGGCCC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| hAT-4b_Ther | Clamp | 571 | 584 | - | 17.89 | GTGAGAGAGAGAGA |
| hAT-4b_Ther | ZNF530 | 362 | 375 | + | 17.44 | GCATGGAGAGGGGA |
| hAT-4b_Ther | eor-1 | 565 | 577 | - | 17.31 | AGAGAGAAAGAGA |
| hAT-4b_Ther | BPC6 | 568 | 588 | + | 16.91 | CTTTCTCTCTCTCTCACACGC |
| hAT-4b_Ther | ZFP42 | 1017 | 1029 | - | 16.71 | CAACATGGCTGCA |
| hAT-4b_Ther | Clamp | 573 | 586 | - | 16.44 | GTGTGAGAGAGAGA |
| hAT-4b_Ther | RAMOSA1 | 573 | 586 | - | 16.41 | GTGTGAGAGAGAGA |
| hAT-4b_Ther | Stat5b | 404 | 412 | + | 16.36 | TTCCCAGAA |
| hAT-4b_Ther | eor-1 | 569 | 581 | - | 16.31 | AGAGAGAGAGAAA |
| hAT-4b_Ther | OVOL1 | 1071 | 1080 | + | 16.02 | ATACCGTTAT |
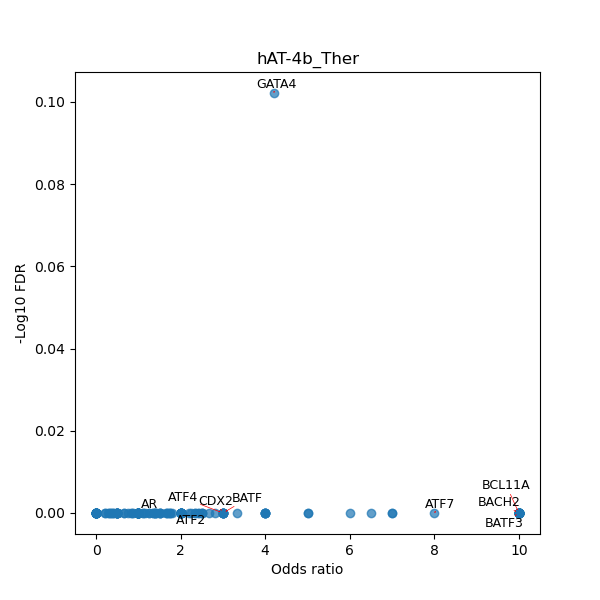
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.