hAT-1_Mam
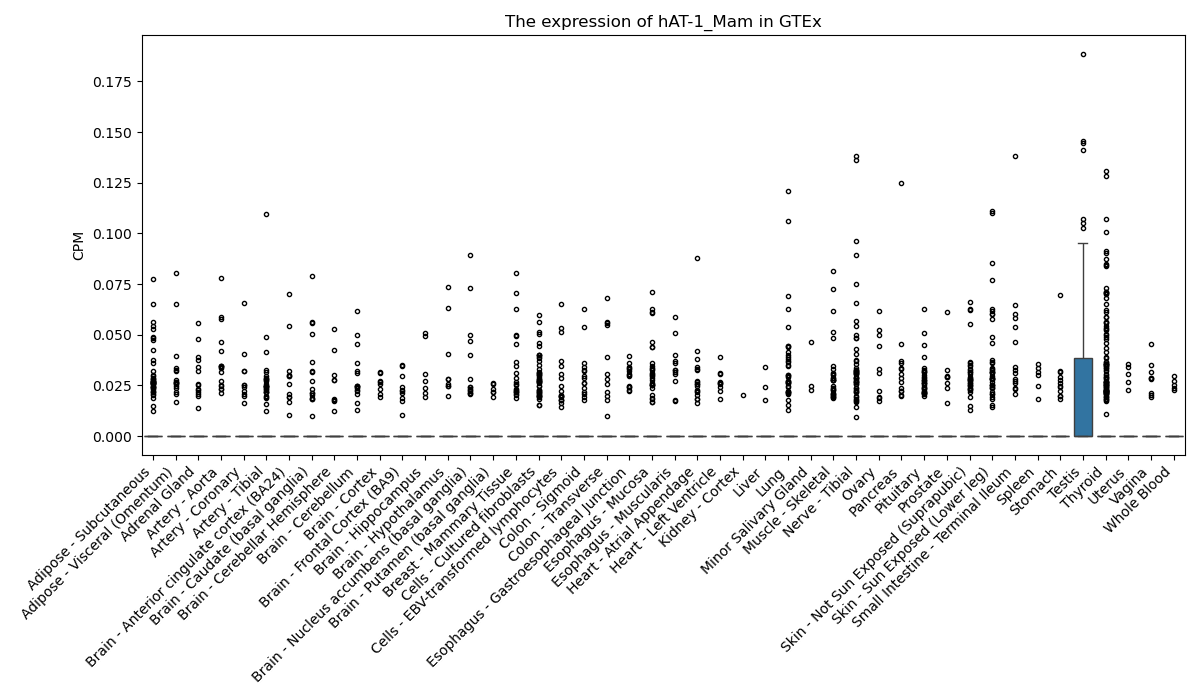
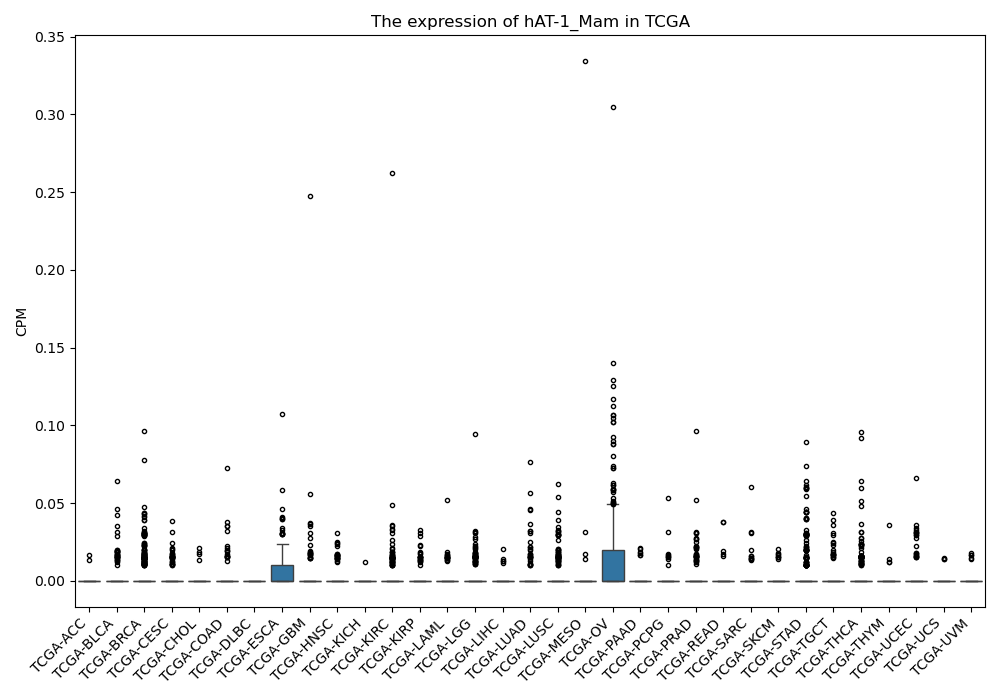
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001293 |
|---|---|
| TE superfamily | Tag1 |
| TE class | DNA |
| Species | Monotremata,Theria_mammals |
| Length | 3469 |
| Kimura value | 32.43 |
| Tau index | 0.0000 |
| Description | hAT-Tag1 DNA transposon, hAT-1_Mam subfamily |
| Comment | 23 bp TIRs. Near-identical to hAT-1_AMi, which was active in the ancestor of all crocodiles. ORF 1007-2917 encodes a transposase related to other Tag1-group hAT tranposases, like hAT-13_AMi_tp and HAT-25_SM_tp. Occasionally found at orthologous places in marsupial and eutheria. There are copies in platypus, but it is unclear that they are at orthologous sites |
| Sequence |
AAATGTAAAGGCCCGGGAAAAAATCACGGAATAATNNGGGAAAAAAATANNCTTTCCCCTTCTTCTCCGAAGAGTTTTTTGCCCTNTTTCCNNNAAAAATCGGAAAAATTGAAAATTTTTNGAAAAATTAGAAAANTTTGGATTATGAANTNTTTTAGAATGATGAATAAAATGTTTAAGATAAGGTGTTCATATATTGCTACCTGGCCTTTACTCCCCCAAAATGATTTCTAAAAACTATGCAATAGGAAGATTTTTATGTAAAACTATCTACAGTATCAGATCATTACAATTCCTGTACACTGAGGACAAGCAGTACAGTACAACTCTCAAATGCAAGAGCAAGATGCATTTCTGCCTCCAGGGGGCACTGCCATTTTCACAAGAGAAGGAAGAAGGATTCAGTGCCTTCTTTGCTTGTGGGCTTCCTCTTGTTGACTTTTGATCTAGAGGTTGGTGGATATGTTCCTGTCCTCCTGACTAGGCCTCAAAGGACCGGGAGTGGAGTGGACACTGGCAGTAGCATCCATTTATTCCTGTAGAGATTAGAAACAAGGAGCTTATTGCCCTGCTTGCTCAAAAGGCTCAGGACAGAGAGAAGTAAGAGCAAGTCTAAAGGTTTTGGAGTTGGAAGGAAGTTGGAACCTGGCTCCTCCTGGCCCTGTGACCTTGAGATCCACAAAGGAGCTGAAAGAAAGTATACTTCTTTATTGAAAGGTAAGAGTTTAAATTTATCTGAATTCAGAATATTAAACTATATTTTAGCTGCATTAGCATATTTTCATAACCTATGTAAATCATAATTGAATTCCTACTAATCCGATGTTTTGTGGCAAAATCACCTACCCAACCTTAGCAGTGAGGTGAGGCCTGCAGGGCTGGAAAGTTGTTTACAGTTAACTCTGAGGAGACGCCATTACTAAATTTGTACATGCTGTAGCTTAGTGTTTATATGCAAGCTAGCCACTAAACTACATTTTTCCTTTTTTCTCCCTTCAGCAAAATGGTGAGACCAACTTCTAGCATATGGAAGCATTTCCACACTGCAAGTTATTCTGAGGGGGGAAAANACGTGATTTGCAAATACTGTCAAATGAAATATGCATTTCCAAATGCTACTAGGATGACCAAACACATCCTTGTATGTAAGAAGTGTCCTGCAGACATTAAAAAATCCTTCATGGGTATAAGTGAGGAGGACCACGATGAAGAAAGTGCAAGTAGCTCTTTCCAGCGTTCTCGATCCAGAAGTCCTCCTGCTGCTGCTTCTGTAACATCAGCATCATCACAAAGGGTTGCCCAATCAACATCCTCGAGTAACCAAATACTGCAAGCAACTGTATACAAAAAACTCTCAATGGCTTCATTTGTAGACAAGATGACACCTGAAGATCAGGAAAAAATTGACCAAGCTCTTGCAAGAGCAATATATTCTTCTGGAACGCCACTGTCAATAACTGAAAATGCATACTGGCAGGAAGTTTTTAAATTACTAAGACCATCATACCGTTTGCCCAGCAGACATTCTTTGAGCAAGCCTCTTTTAGAGTCAGAATATGAACGGGTAATGGAATCTGTGCAAGGAAGAATCAATGAAGCACTGTGTCTTGCAGTACTTACAGATGGATGGACAAATGTGCGGGGAGAAGGAATCATAAACTTTGTGATAACAACACCTCAACCAGTTTTTTACAGAAGCATTGAAACAGGAGAGAACAGACACACTGCAGAGTATATCAGTTGTAAAATATGTGAAGTCCTACAGAAGACAGGGAGCGGTAAAGTATTTGCTTTGCTGACTGACAATGCAAGTAACATGAAAGCAGCATGGGAGATCATAACGGATAAATANCCACATATTACTGCAATAGGATGTGCATCCCATGGCCTAAACTTGCTTCTTAATGACATCATGAAGTTGGACACTCTCCAAAAGATCTANAGAAGAGCAAAAGAAGTTATAAAACATGTAAAAGCTACTCATATTGTAGCTGCTGTCTTCAAGAAGAAGCAAGAAGAAAAGAATGCAAAGAATAAAATAGTGACACTGAAACTCTGTAGTAAAACTCGATGGGGTGGAGTGGTAATGTCCTTTGAGAGTTTACTAAAAAACAAAGAAGCTCTTCAAGAAACAGTAATAGTTGAGGATCTTAAAGTACCCAGAAGTGTAAGGAACACTGTACTTGATGAGGATGTCTTCTGGGTTCAACTGCAGAACTCCCTGAAGATTCTAAAGCCAATTTCTTCAGCAATCACTGCATCTGAATCAGACAGTGCACTTTTGTCAGACATTCCTTACCAAATGGCCCAGATAAAATCAACTGTTTTTGAGAATCTAAGTGTATTTCCGCTTACAAGTACAGAAGAAAGTAAAGTGAAAGACTTTATTAAGAAACGGGAAGAATTTTGTTGTCAGCCAATTCATGCTGCTGCAAATCTCTTGGATCCAAGATTCAAAGGCAGAAATGTAAGTGATGATAGCACTGCTACTGCATTTGACTGGATTACAAAACAAGCAGCACACTTAGGACTTGATGTTGGAAAGGTACTTTCAAATGTTGCAGAATACAGAACATCTTCAGGTGTTTGGTCCAGGAATGCAGTCTGGGATTCTGCCAAGCATATCCCTCCTGCAACATGGTGGCAAGGCCTTTGCACGACACAGCCTCTGACTCCTCTTGCTTCTCGCCTGCTTCAGATTCCTCCATCTTCTGCAGCATGTGAAAGAAATTGGTCTATGTTTGGAAATACTCGCACCAAATCAAGAAATAAACTAACATGTGAAAGAGTAGAGAAGCTTGTCTCAATCCGGGCAAACCTTCAGTTTTCAGAAGTCCGTGAAGATGATAAAAATGAAGCATCAGAAGAAGCAGAAACTGCTGATAGCGATAGCTCAGAAAATGAGACTGATTAGACAAAGTTCCATGTGGCAAAAGTTGTATTCAATGAGTTTGGGTTTTTTTCCTTTTTTCCCTCAGCTCCTTTTCTGAAAATGAGTGCTTTCTGTATCTGCTTGACACTGTGGAGGACACTAAGTCCAGAAAACATAATAACTTTCTTTGTTTTACTATAATGTTGATGGTTTCTTCTGTTCTCAAGTTTTTGCCTGCTTCTCATAAACTGGCAAAAACTAAGAGATGCTAGATTCCTTAAAGTTCAGCAGCTTGTAGCTCCATTTGTAACAAAAATTAAAAACATTTAAAAGTAAATTCAATTAGTAAATTTGTAATTATTGTCTGAATAAACTATACTTTTAAATTTAATATACCAAACATGATAATTTTATGAGCAAACCAAGTTATACATAATACATTTTTATGTTCCTAAACTAACAAAGTGACTTTAGATCTGTAAAGGGCGCTAAAAAAATAAGTATTGCATATATTTCAATTTTCCCCAAAATTTCCATTTTTTCCCGAAAAACGTTTTCTCCCATGGCTTCAAAATTTCCGGAAATTTTACATCTCTA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| hAT-1_Mam | CTCF | 357 | 371 | + | 20.52 | GCCTCCAGGGGGCAC |
| hAT-1_Mam | ZNF547 | 764 | 776 | - | 19.76 | TGCTAATGCAGCT |
| hAT-1_Mam | CTCF | 340 | 370 | + | 17.91 | GAGCAAGATGCATTTCTGCCTCCAGGGGGCA |
| hAT-1_Mam | GFI1 | 2250 | 2260 | + | 17.74 | CAATCACTGCA |
| hAT-1_Mam | WRKY71 | 434 | 443 | - | 17.48 | AAAAGTCAAC |
| hAT-1_Mam | ZmbZIP57 | 1905 | 1914 | - | 17.44 | ATGATGTCAT |
| hAT-1_Mam | WRKY8 | 433 | 443 | - | 17.43 | AAAAGTCAACA |
| hAT-1_Mam | WRKY45 | 434 | 443 | - | 17.42 | AAAAGTCAAC |
| hAT-1_Mam | JUN | 1905 | 1914 | - | 17.08 | ATGATGTCAT |
| hAT-1_Mam | ZNF707 | 493 | 507 | - | 16.91 | CTCCACTCCCGGTCC |
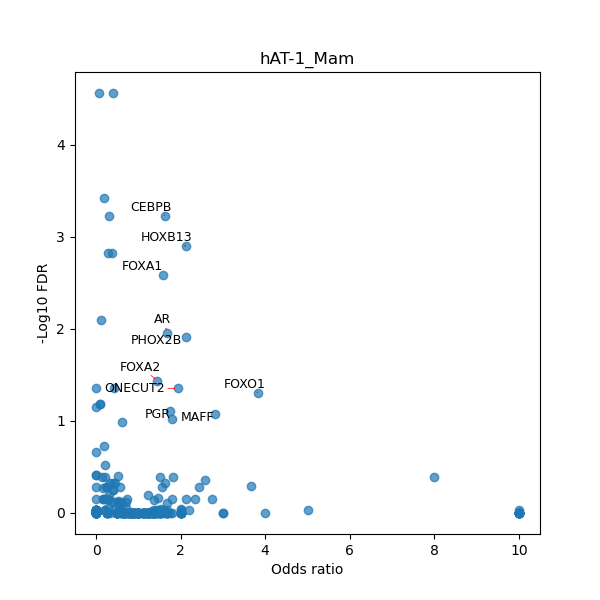
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.