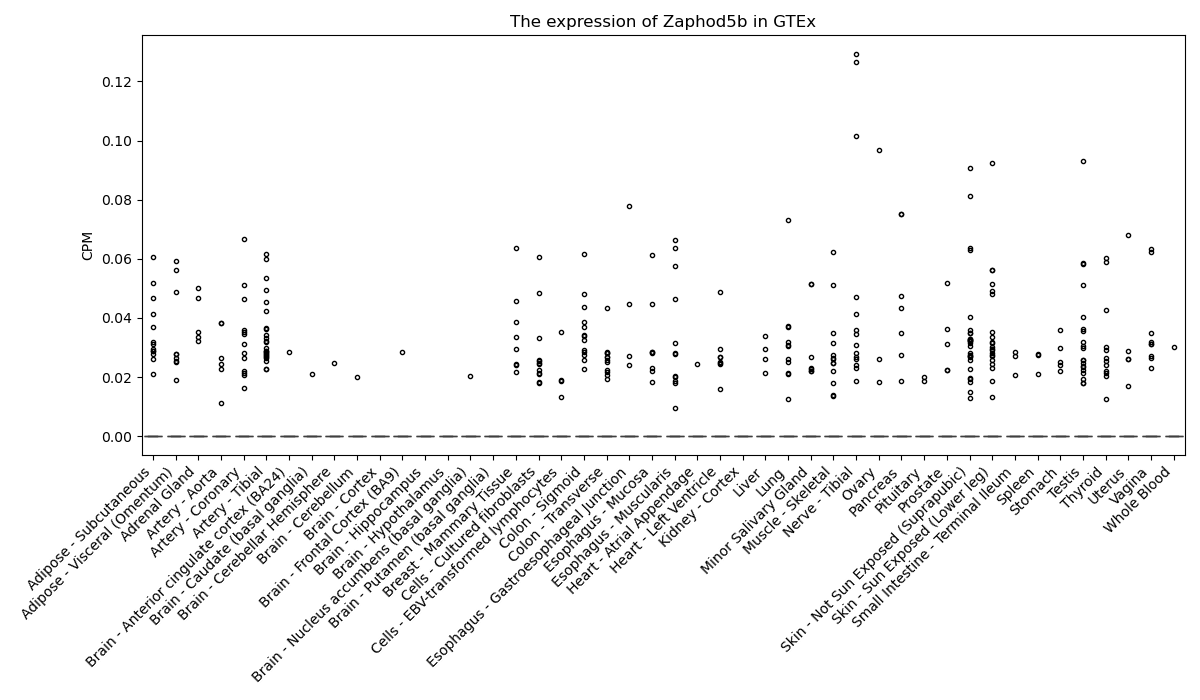
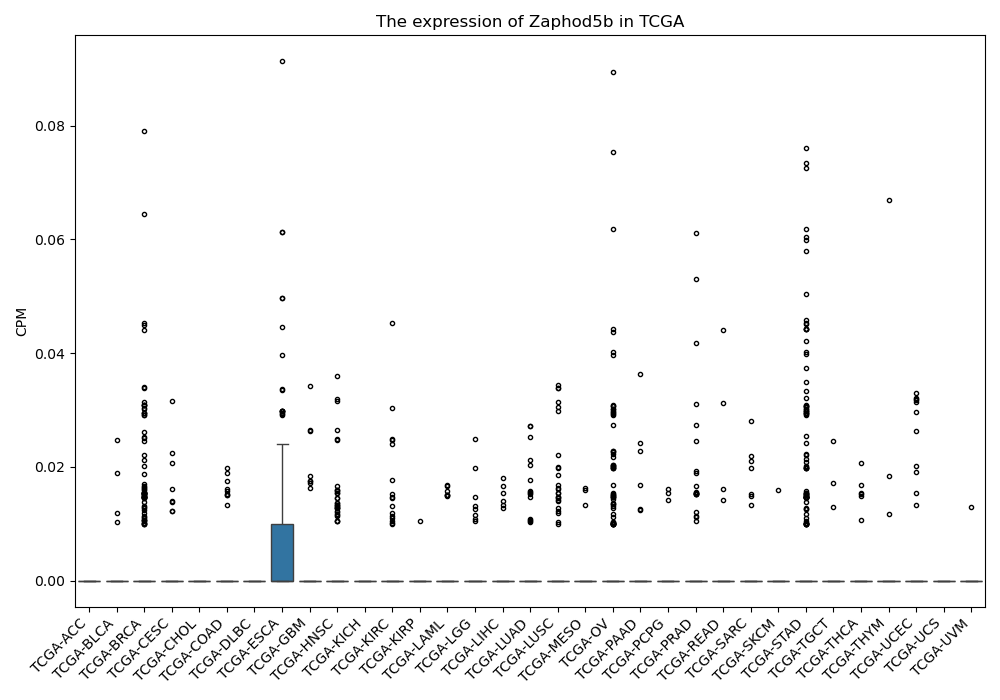
Zaphod5b
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001192 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 406 |
| Kimura value | 27.74 |
| Tau index | 0.0000 |
| Description | hAT-Tip100 DNA transposon, Zaphod5b subfamily |
| Comment | 8 bp TSD (hard to see). 17 bp TIRs. Present Eutheria, absent in marsupials. |
| Sequence |
CAGAGGCNTAACAACTACAGTACGAGCCCGTGTGCAACTCAGGAAGTGGGCCCCCTCTCCCTCCTCTCCCCCCCTCTNCCTCCGTCTTACGTCAAACATTTGCTTCGCTTAAAGCTCGTAAAATGCCATTTCATTAAATCGTGTTTTCATTTATGTAAGATGACTGTCATGCCAAAGCTATTTTATTGTTATAGAAGTTAGGAAAATATAAAGAAAATACATTTTTAGAATAAAATAACACAAAACATATTTTAGTTCTATCTTCTTGATTTTTAAATGAAATTATGGATCTCCAAGCCTAATTTTTGTTTCATGNCAATTAAAATATTTCCTCCTGGGGCCCTCTCTGGGCCCCCTTGCCCCTGGACCANNTNTCAGCTGCACACTCTNCAGCTTTGATGGTTAC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Zaphod5b | HOXC12 | 114 | 123 | + | 14.80 | GCTCGTAAAA |
| Zaphod5b | Glyma19g26560.1 | 46 | 53 | - | 14.75 | GGGCCCAC |
| Zaphod5b | GLYMA19G26560 | 46 | 53 | - | 14.75 | GGGCCCAC |
| Zaphod5b | ZNF692 | 347 | 354 | - | 14.58 | GGGCCCAG |
| Zaphod5b | TCP16 | 46 | 53 | + | 14.51 | GTGGGCCC |
| Zaphod5b | TCP22 | 46 | 56 | + | 14.42 | GTGGGCCCCCT |
| Zaphod5b | Erg | 39 | 48 | + | 14.36 | TCAGGAAGTG |
| Zaphod5b | GABPA | 39 | 48 | - | 14.34 | CACTTCCTGA |
| Zaphod5b | ZNF148 | 52 | 61 | + | 14.28 | CCCCTCTCCC |
| Zaphod5b | ZNF574 | 339 | 352 | - | 14.22 | GCCCAGAGAGGGCC |
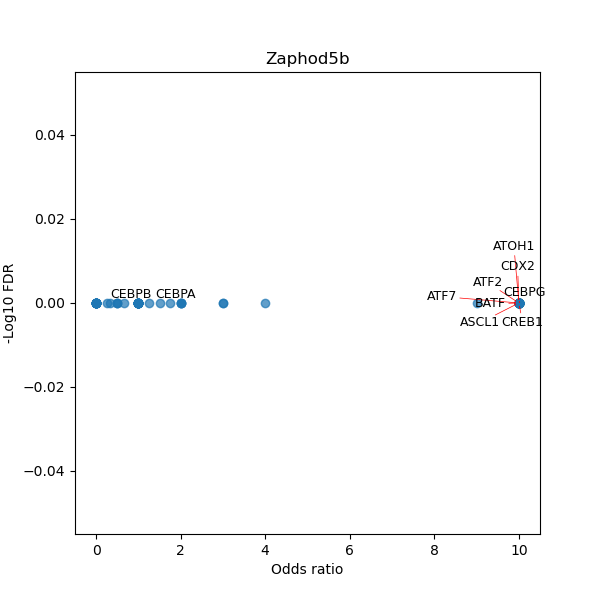
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.