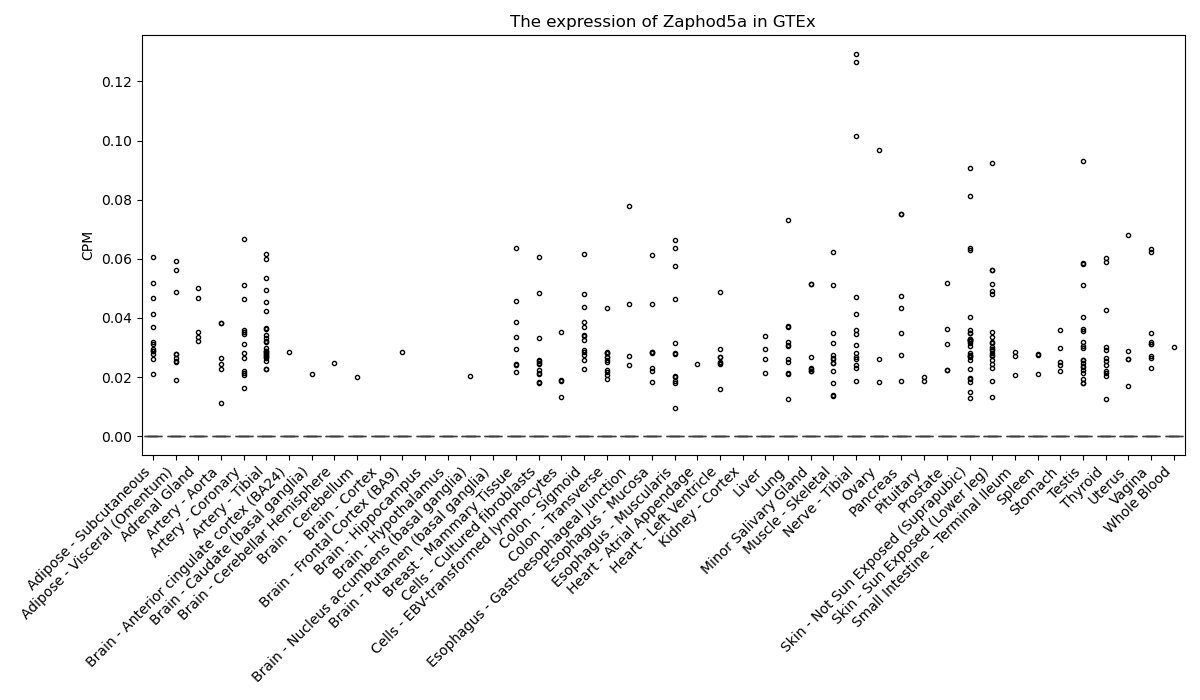
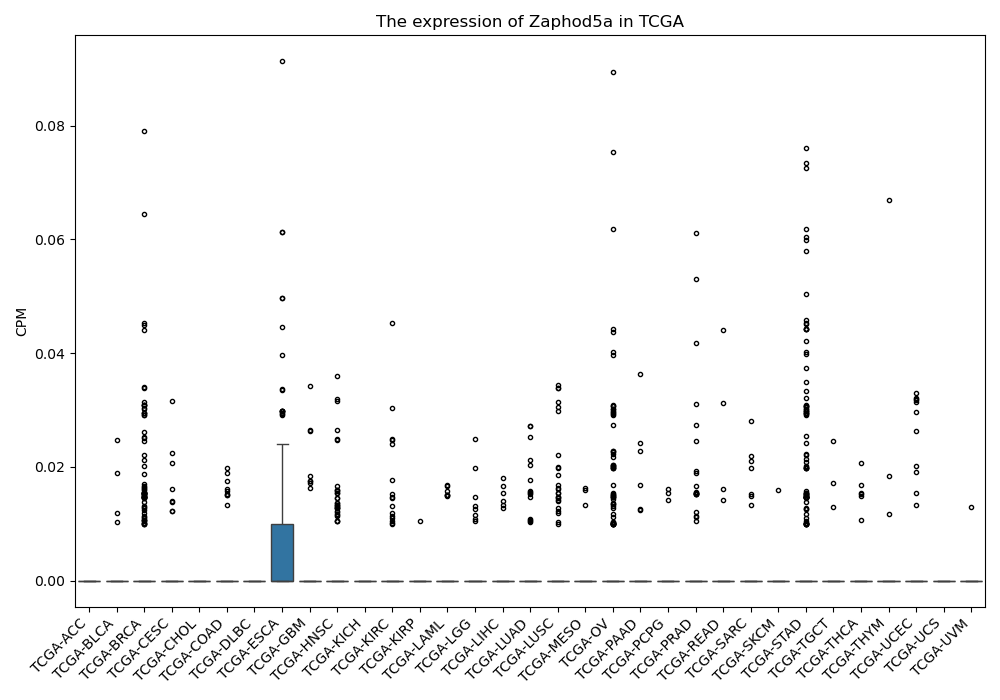
Zaphod5a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001254 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 504 |
| Kimura value | 27.61 |
| Tau index | 0.0000 |
| Description | hAT-Tip100 DNA transposon, Zaphod5a subfamily |
| Comment | 8 bp TSD (hard to see). TIRs distantly shared with family members DNA8-60_AP in the pea aphid and hAT-N1001d3_Croc in ancient crocodilians. Pos 127-222 is the remainder of the coding region, matching the C-terminus of Zaphod and Arthur transposases. Present in placentals, absent from marsupials. |
| Sequence |
CAGGGGCATAACAACCACAGTGTGGGCCCGCGTGCAACTCAAGAAGTGGGCCCCCTCTCCCTCCGCCTCATGTCTCTTCAAAAACAAATCTCAGTGCACAGTGCAGTGAAACACGAAACGAAACAGAGCTCGCTTACTATTGAAAGCAAAAGAGCACGGCAACTTAATTTAAACAAGATAATTCAAACATTTGCTTCGCTTAAAGCTCGTAAAATGCCATTTCATTAAATCGTGTTTTCATTTATGTAAGATGACTGTCATGCCAAAGCTATTTTATTGTTATAGAAGTTAGGAAAATATAAAGAAAATACATTTTTAGAATAAAATAACACAAAACATATTTTAGTTCTATCTTCTTGATTTTTAAATGAAATTATGGATCTCCAAGCCTAATTTTTGTTTCATGTCAATTAAAACATTTCCTCCTGGGGCCCTCTATGGGCCCNCTTGCCCCTGGACCGTCTGNCAGCTGCACACTCTGCGCCCCTGGTAGTTACGCTCTTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Zaphod5a | OsI_08196 | 46 | 53 | - | 15.69 | GGGCCCAC |
| Zaphod5a | PLAGL2 | 48 | 55 | + | 15.67 | GGGCCCCC |
| Zaphod5a | ARALYDRAFT_493022 | 22 | 29 | - | 15.63 | GGGCCCAC |
| Zaphod5a | ARALYDRAFT_493022 | 46 | 53 | - | 15.63 | GGGCCCAC |
| Zaphod5a | CG7928 | 480 | 491 | + | 15.54 | TGCGCCCCTGGT |
| Zaphod5a | ARALYDRAFT_495258 | 22 | 29 | - | 15.43 | GGGCCCAC |
| Zaphod5a | ARALYDRAFT_495258 | 46 | 53 | - | 15.43 | GGGCCCAC |
| Zaphod5a | ARALYDRAFT_484486 | 22 | 29 | - | 15.43 | GGGCCCAC |
| Zaphod5a | ARALYDRAFT_484486 | 46 | 53 | - | 15.43 | GGGCCCAC |
| Zaphod5a | REF6 | 120 | 128 | + | 15.41 | GAAACAGAG |
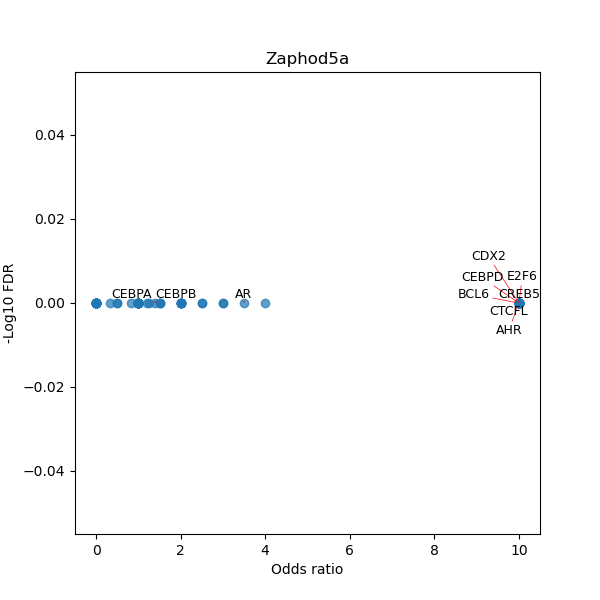
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.