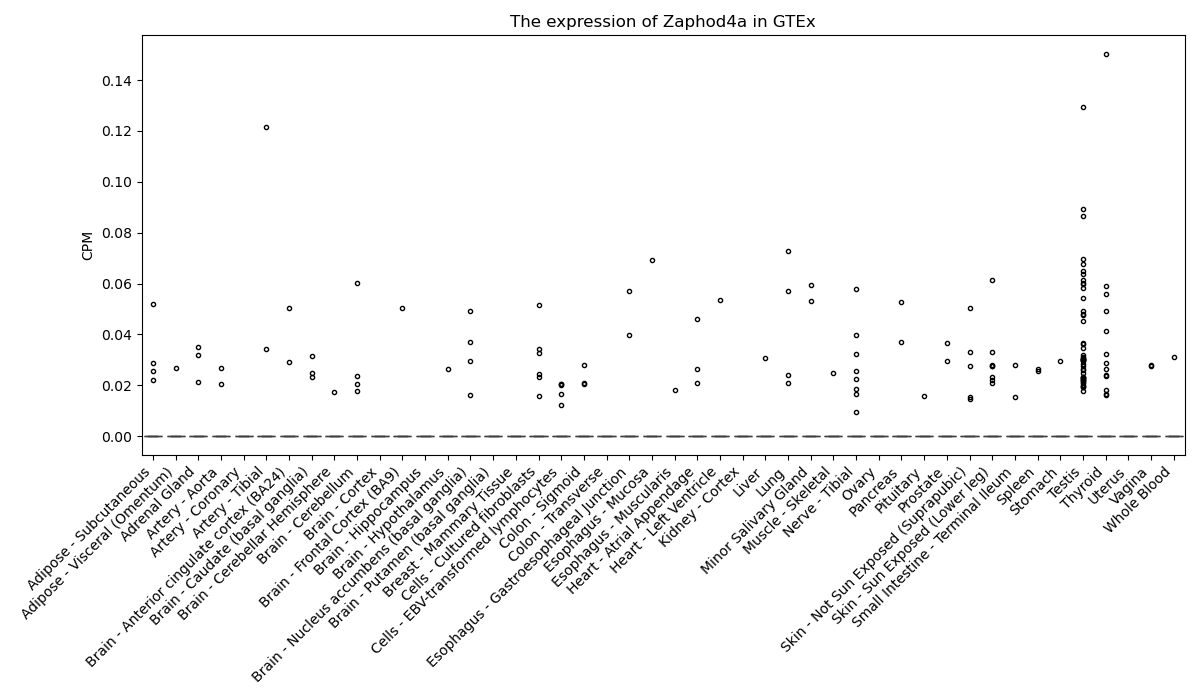
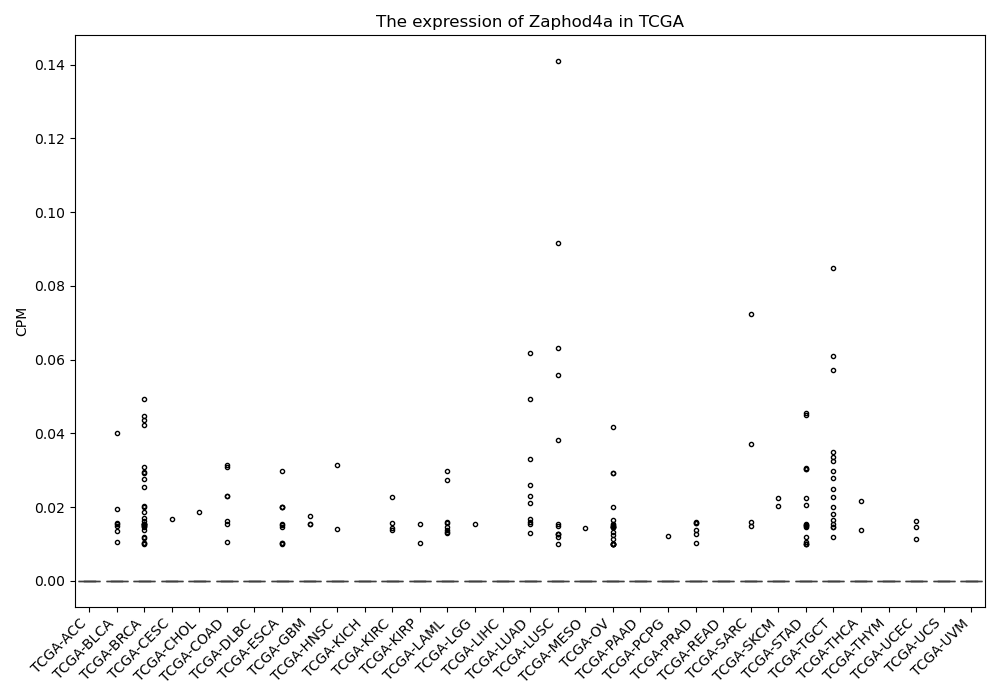
Zaphod4a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001290 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 1823 |
| Kimura value | 29.46 |
| Tau index | 0.0000 |
| Description | hAT-Tip100 DNA transposon, Zaphod4a subfamily |
| Comment | 8 bp TSDs. Only 7 bp TIRs. Non-autonomous. Remaining ORF 1022-1567 encodes the C-terminal quarter of a transposase closest to Arthur1_Cin_tp and Arthur3_Api_tp, but closer to original Zaphod than to original Arthur. Sloppy hairpin structure at 637-736. Pos 303-338 is inverse duplicated at pos 576-541. Present at orthologous sites in elephant and humans, absent in opossum. |
| Sequence |
CAGTGGCGAAACTAAGCCGATGCAGCCGATGCGTTGCATCGGGGCCCATGGGCTATAAGGGGCCCATGTGAGTTGAAAGTTTCAATAGTATTGATCGTGGCCACTGAATAGGCTAATGCATTTAAAAATCAGTATGACATCATTAGCAAACAAACGAAATTGTACTTGAATAGAATTCAACTTTGGCTGGATTTTCTTAAAAATCTGGCTACAGTATTATGGCGCTGGCGCACATATTTTAATTTGGCGCTGGGCCTCCATGACAATAGAGTAGGTGAAATTCCATAAATGTTCGAGCGCATCTATACTCTATTCCAATGACCTACGGAGTAATAAATCATTCTGTCAGTTGCGCAGTAACATTTAAGTCCCACCCTCCTAATCCCTAATTGGCTAAGCAGTTGCAAGACCAAATTACATAGTACTGAAGAAAATATAAACACGAGCATGTAAACAATATCACAATAGATCACTGTACTTGTCAAATTTACCCTGCGTAATTCAGACTTTCTCTATTTCGCCAAAGAACGCCTCTCGCGCATTTATTACTCCGCAGGTCATTGGAATAGAGTATAGCACGCCTGGATTATTATAATGTGCAAAGAGATCGTACATATATCTCGCCGGTACGTCGGGAGCCCCTATGAAAGCTCTTTGATCTTTGCCGGCTTTTTAGCCGAAAGGATTCGGAAGCCTTTGTCGGCATCGGCAAAGACAAAGAGCTGTCGCAGCGGTTGATAGCCCTTTGATATTTGGCCGCTTTTTAGCCGAAAGGATGCGGAAGTCTTTGTCGGAATCATGTATTAATATATAGTGGACAATGCCATTGTCTTAAAGGCTAGGTGAAAATTGATTTTTTAATCATGTTTCCCGCATCGAAAAACTCCCTTGGCCGAAATTTCGCCGAGATCCGACGATATTTAAAANATTTTTGCATCGTCTGCTTCGGCCTGTCGATATGCCGAGAGCCAATATGATAGCTCTCTGATCTTCGGCAGTTTTTTAGGCGAAGGGATTATAGTTCATCATTTTTCATCGTGTAATTGATATAGTAACTACCCAGCTAGACACGAGATTTCAAGGGCTAAGTACAGTTGCTGACTTGTTTGCATTTCTTACTCCAACCCAATTAATTCAAATGGATGAAAAGTCCATTGTGAAAAATGCACAGAAGGTGCAAAAGAAATTTTCAAGTGACCTATCAGAATCATTACCAATGCAATTACTGCTTTGCGTAAAATCATTAAAAAGTGAAATTGAAAAAATTAAAACTATCTGCGAATTTGCCGAGATAGTTATTATAAAATACTACGACATGAGTGCAAGTTTTTCTGAAGTGTGTTCTTTATTAATTTTATTTTTGACAATTCCCGTCACTGTATCTATAGCAGAACGTTCTTTCTCCAAATTAAAACTAATTAAAAACTATTTAAGGAACACTATGGGACAACAACGTTTAACAGAGTTAGCACTACTCAGCATAGAAGCTCAAGCAGCTGAAAAAATGGAAGTTGCAAAACTTATCGATGATTTTGCTAATGCCAAATCACGACGAAGACCATTCTAAGTTTGCTTAAAATTTTTAAATTTGTTTATATTGACAGAATATTGTCTTTGTTTATTATATTGTCATGTTTTGCTTTAGATTTTAAGTTTCAAAGTTTGTTTATATAGTAATTTTTTTGTTTTATCAAAAAGAATGCTATTTGGTTATTAATAAAGTTATGTACCTATGTATGGCAAAATAAGCTCTTTTTGTTGTACGTAGGGGAGGCCCACATTAGACTTTGCATCGGGGCCCATTTTTACTAGTTATGCCACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Zaphod4a | lsy-2 | 209 | 216 | + | 15.13 | CTACAGTA |
| Zaphod4a | ZHD1 | 1414 | 1422 | + | 15.13 | ACTAATTAA |
| Zaphod4a | TCF7L2 | 653 | 661 | - | 14.98 | AGATCAAAG |
| Zaphod4a | EREB127 | 698 | 705 | - | 14.97 | TGCCGACA |
| Zaphod4a | ARF29 | 697 | 704 | - | 14.91 | GCCGACAA |
| Zaphod4a | DOF3.4 | 1246 | 1262 | - | 14.82 | TTTCAATTTCACTTTTT |
| Zaphod4a | MYB43 | 268 | 279 | - | 14.80 | TTCACCTACTCT |
| Zaphod4a | DREB1G | 698 | 705 | - | 14.74 | TGCCGACA |
| Zaphod4a | sens | 1544 | 1553 | + | 14.74 | AAATCACGAC |
| Zaphod4a | ZmbZIP25 | 133 | 142 | + | 14.73 | TATGACATCA |
TFBS enrichment in GRCh38
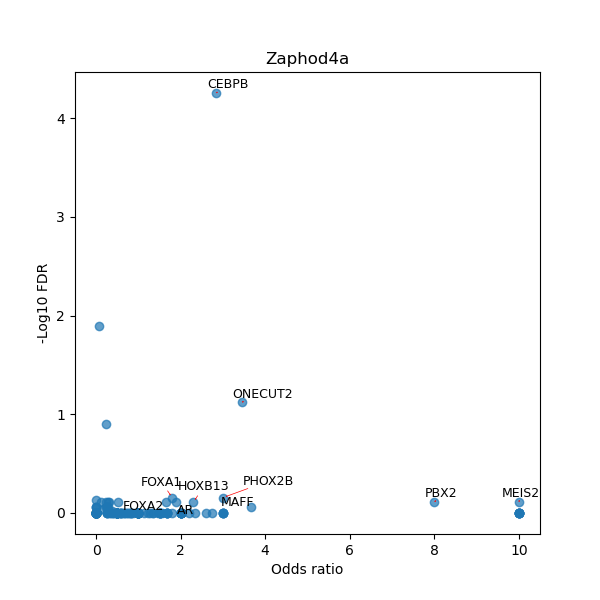
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.