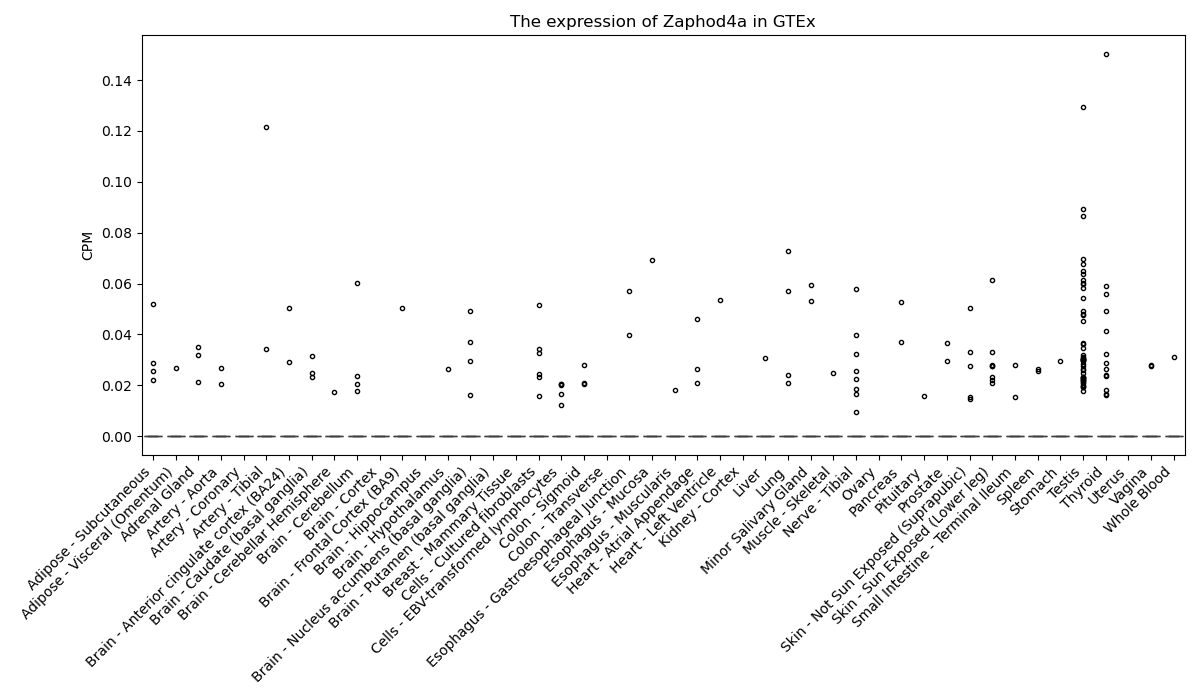
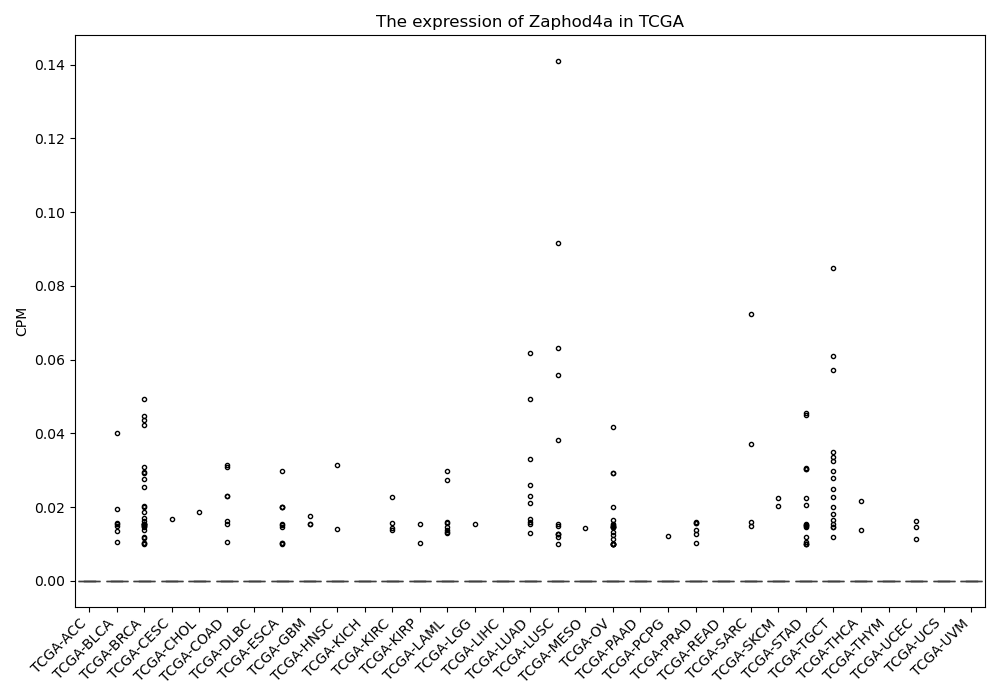
Zaphod4a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001290 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 1823 |
| Kimura value | 29.46 |
| Tau index | 0.0000 |
| Description | hAT-Tip100 DNA transposon, Zaphod4a subfamily |
| Comment | 8 bp TSDs. Only 7 bp TIRs. Non-autonomous. Remaining ORF 1022-1567 encodes the C-terminal quarter of a transposase closest to Arthur1_Cin_tp and Arthur3_Api_tp, but closer to original Zaphod than to original Arthur. Sloppy hairpin structure at 637-736. Pos 303-338 is inverse duplicated at pos 576-541. Present at orthologous sites in elephant and humans, absent in opossum. |
| Sequence |
CAGTGGCGAAACTAAGCCGATGCAGCCGATGCGTTGCATCGGGGCCCATGGGCTATAAGGGGCCCATGTGAGTTGAAAGTTTCAATAGTATTGATCGTGGCCACTGAATAGGCTAATGCATTTAAAAATCAGTATGACATCATTAGCAAACAAACGAAATTGTACTTGAATAGAATTCAACTTTGGCTGGATTTTCTTAAAAATCTGGCTACAGTATTATGGCGCTGGCGCACATATTTTAATTTGGCGCTGGGCCTCCATGACAATAGAGTAGGTGAAATTCCATAAATGTTCGAGCGCATCTATACTCTATTCCAATGACCTACGGAGTAATAAATCATTCTGTCAGTTGCGCAGTAACATTTAAGTCCCACCCTCCTAATCCCTAATTGGCTAAGCAGTTGCAAGACCAAATTACATAGTACTGAAGAAAATATAAACACGAGCATGTAAACAATATCACAATAGATCACTGTACTTGTCAAATTTACCCTGCGTAATTCAGACTTTCTCTATTTCGCCAAAGAACGCCTCTCGCGCATTTATTACTCCGCAGGTCATTGGAATAGAGTATAGCACGCCTGGATTATTATAATGTGCAAAGAGATCGTACATATATCTCGCCGGTACGTCGGGAGCCCCTATGAAAGCTCTTTGATCTTTGCCGGCTTTTTAGCCGAAAGGATTCGGAAGCCTTTGTCGGCATCGGCAAAGACAAAGAGCTGTCGCAGCGGTTGATAGCCCTTTGATATTTGGCCGCTTTTTAGCCGAAAGGATGCGGAAGTCTTTGTCGGAATCATGTATTAATATATAGTGGACAATGCCATTGTCTTAAAGGCTAGGTGAAAATTGATTTTTTAATCATGTTTCCCGCATCGAAAAACTCCCTTGGCCGAAATTTCGCCGAGATCCGACGATATTTAAAANATTTTTGCATCGTCTGCTTCGGCCTGTCGATATGCCGAGAGCCAATATGATAGCTCTCTGATCTTCGGCAGTTTTTTAGGCGAAGGGATTATAGTTCATCATTTTTCATCGTGTAATTGATATAGTAACTACCCAGCTAGACACGAGATTTCAAGGGCTAAGTACAGTTGCTGACTTGTTTGCATTTCTTACTCCAACCCAATTAATTCAAATGGATGAAAAGTCCATTGTGAAAAATGCACAGAAGGTGCAAAAGAAATTTTCAAGTGACCTATCAGAATCATTACCAATGCAATTACTGCTTTGCGTAAAATCATTAAAAAGTGAAATTGAAAAAATTAAAACTATCTGCGAATTTGCCGAGATAGTTATTATAAAATACTACGACATGAGTGCAAGTTTTTCTGAAGTGTGTTCTTTATTAATTTTATTTTTGACAATTCCCGTCACTGTATCTATAGCAGAACGTTCTTTCTCCAAATTAAAACTAATTAAAAACTATTTAAGGAACACTATGGGACAACAACGTTTAACAGAGTTAGCACTACTCAGCATAGAAGCTCAAGCAGCTGAAAAAATGGAAGTTGCAAAACTTATCGATGATTTTGCTAATGCCAAATCACGACGAAGACCATTCTAAGTTTGCTTAAAATTTTTAAATTTGTTTATATTGACAGAATATTGTCTTTGTTTATTATATTGTCATGTTTTGCTTTAGATTTTAAGTTTCAAAGTTTGTTTATATAGTAATTTTTTTGTTTTATCAAAAAGAATGCTATTTGGTTATTAATAAAGTTATGTACCTATGTATGGCAAAATAAGCTCTTTTTGTTGTACGTAGGGGAGGCCCACATTAGACTTTGCATCGGGGCCCATTTTTACTAGTTATGCCACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Zaphod4a | CHES-1-like | 449 | 458 | + | 15.90 | TGTAAACAAT |
| Zaphod4a | TSO1 | 1574 | 1588 | - | 15.89 | ATTTAAAAATTTTAA |
| Zaphod4a | TSO1 | 1582 | 1596 | - | 15.81 | ATAAACAAATTTAAA |
| Zaphod4a | SOL1 | 1574 | 1588 | + | 15.33 | TTAAAATTTTTAAAT |
| Zaphod4a | STB3 | 1256 | 1266 | - | 15.30 | ATTTTTTCAAT |
| Zaphod4a | SOL1 | 1581 | 1595 | - | 15.22 | TAAACAAATTTAAAA |
| Zaphod4a | FOXN3 | 450 | 457 | + | 15.19 | GTAAACAA |
| Zaphod4a | Sox6 | 824 | 833 | + | 15.16 | CCATTGTCTT |
| Zaphod4a | CHES-1-like | 1588 | 1597 | - | 15.15 | TATAAACAAA |
| Zaphod4a | CHES-1-like | 1662 | 1671 | - | 15.15 | TATAAACAAA |
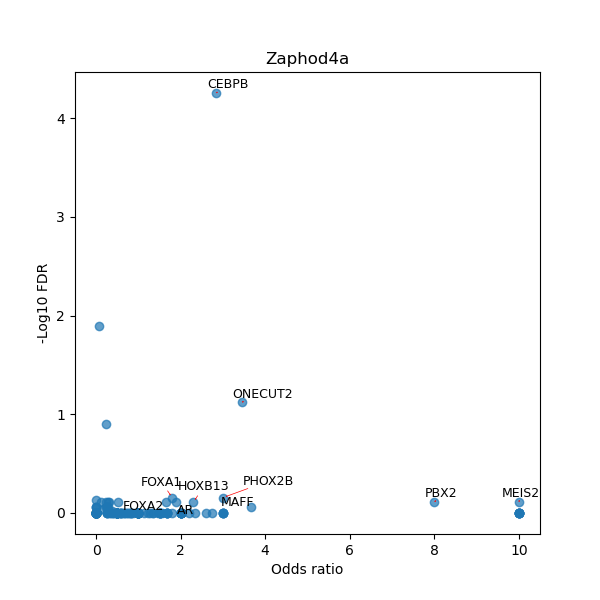
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.