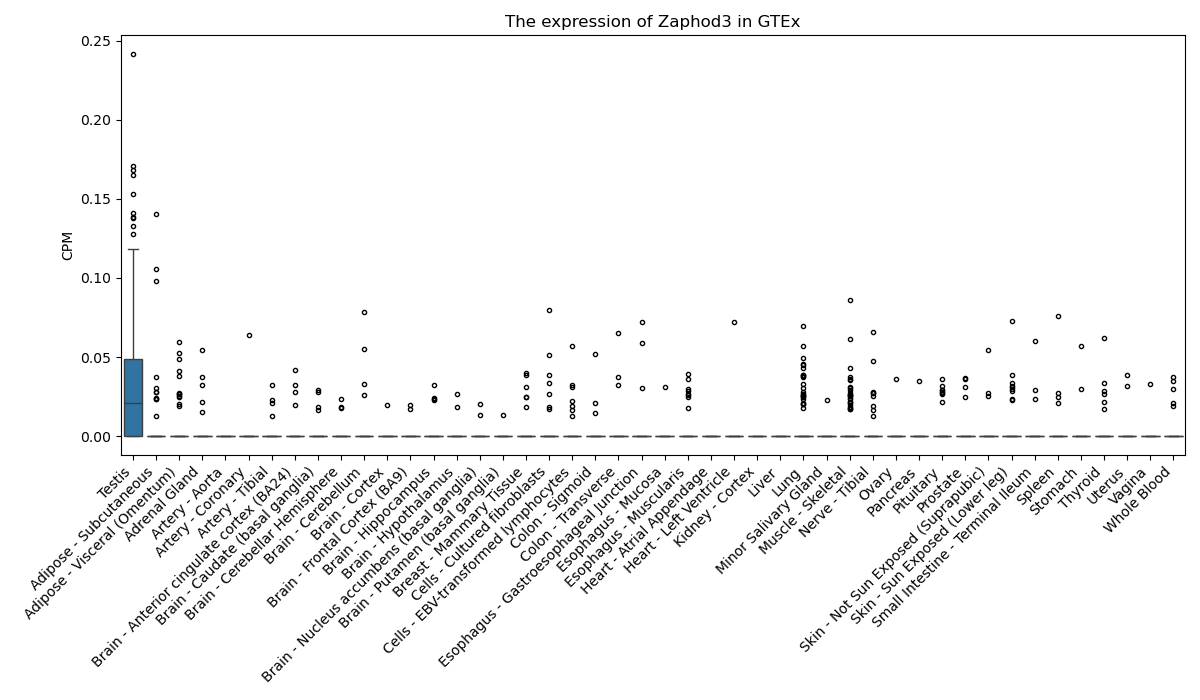
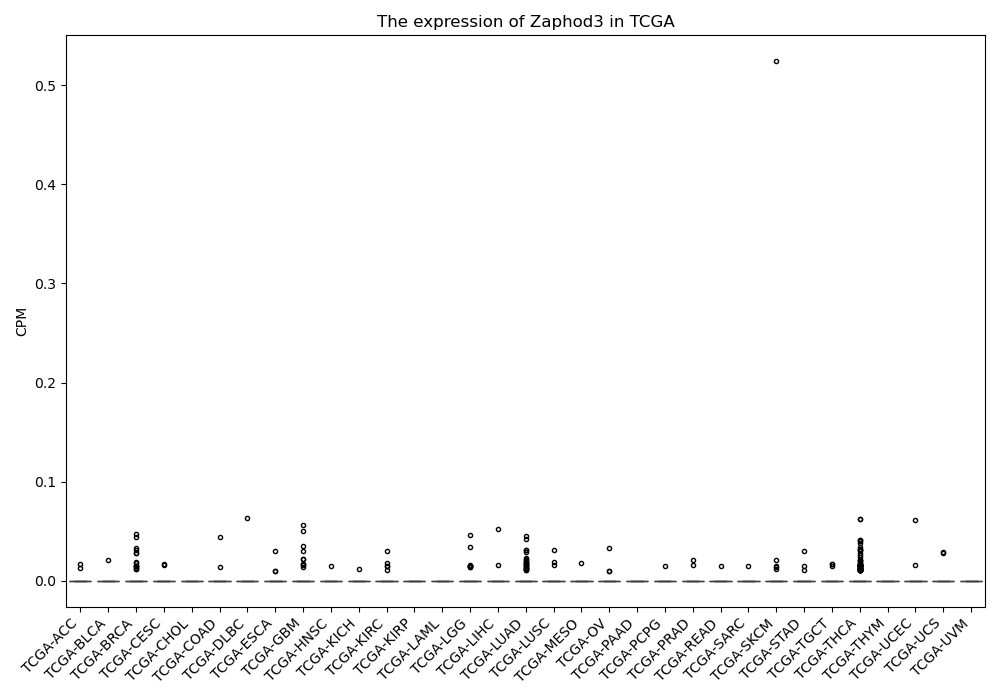
Zaphod3
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001125 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 2624 |
| Kimura value | 22.08 |
| Tau index | 1.0000 |
| Description | hAT-Tip100 DNA transposon, Zaphod3 subfamily |
| Comment | Zaphod3 encodes an ORF (~821..2293) that is roughly equally similar to that of Zaphod and Arthur. MER97a-d are non-autonomous, internal deletions of Zaphod3. |
| Sequence |
CAGTGGCGTACCAAGGGCGGGGCGGTGGGAGCGGTCCGCCCCGGGTGCAGGCAATAAGGGGGTGCATTGTCTGTAGAGAATTTAAAAACAATAATAAAACCGACTAAAAGTCGGTCTGCTTTTTATTATCACCATGCGCCGGCAATTCTAAACAATGTCAGTGATAAAATACTCCTCCCCGAAAAATCTTTTGTTGGTCTAAGTTCTAAACAATTGCTGCGGTTACTGTTGAGTTTTAATAATATATATGTAAGCTTCAAATTAGCACATTTTTATTACTTATCCTTTAATAAACATTGTATTCTACATGGAAGTTAATTCGGAGAACTCCCAGTTATACAGTCGGCCCCCGACACACGCGGACTCAGCTACACGCGTTCGTTTCGAGAGTAAGTTCGTAACGGTTCGGAATCGTTCGAGCTCGCTTCGGGCGCAGTTCGTGTCTCCAACCCCTGTGGTACTACATATTCCTGCGTTTAAACAGTAGATTCGAAATAAACAATGATAGCACAGTGATTGTAAAGACGAAGAAACAGAACTTGAGTTACTTCAATTCTGTCATTCTATGTGACCACTTGGAGTTTTTATTTGTGTTTAAAATTTAAAACAGTGAAACAGAGTGCGAACTGCGAGGTGTAATATTTTTGTTTGGTAAGTGCAAATTTTAGTTCATACATGAAATATTTTACTGAATTTGAATAATATCTTTAAAATTGAAATTTATTCTTTTTAAATTGTTAATTGTTTTAAAACTAAAGAATGAATCAAGAAAATAAAACATTACATCAGTGGTACGATTTAGTAGTGGCAGATACTGAAAGCANAATGCCTNGGAATTCTTTCTGNTAGCACTCTNGATGCTTCGAACACTGATCAAATGTGCGAAGTGGTCAGATATGNCNATATTGAAGATGGTAAAGTTGAAATAAAGGAATCATTCTTGGGCTTCTTTCCTATTTCCGGGAAGACTGNTGCTGAACTTNCAGAAGAGANNTCGAAACAACTGGACGGTGATGGGCGGGACATAANCNTCTNCTGTGGNCAAGGATATAACGATGCTGCANNTGCGGCCGGTATTCACGGTGGNGTTCANGCAAAAATCGAAGANATTAATCCCAAATCNTTATTTNTATCTTGTGCAAATCATTCTCTGAACCTTTGCGGAGTTCACTCTTTTGGAAGTATTTCTTCATGCGTGACGTTTTTTNGAACTTTGGAAAAAAATTATTCATTCTTTTCAGTCTCACCTCATCGATGGAAAACGCTGCAGAATGTAGGTATAACAGTGAAAAGACTTTCCCAGACAAGATGGAATGCTCANTATGAAGCTGTGCGCGCAGTANAGACAAATTTTGAAAAGTTAATCTCAACCTTTGAAGTACTGTGTAATCCAAAAGAAAATGTGGACNCAAGAGGATCNGCTCAGATTTTNCTCTCTGCTGTATGCGATTTTTCTTTTCTGAGTTATCTTTTTTTCTGGTGTGAAGTTTTAGATGAGGTTAATCAGACACAAAAATATTTGCAAACAGCCAGAATCAGCCTTGAACAATGTACAGTGAAACACCAAGCTTTAAAATTGTTCCTTGAAGATCGGCGCACAGAAATTGTGGAGAAGGCCATTAACTATGCAACAACAAAATGTAAGGAAATGGACATTTACATAGAAAAAAGAATCAAATTTCGAAGAAGAATGCCAGGAGAAACGACAAAAGATGCTGGTCTTACATTGCCAGAAGAAATCAAAAGGGCAATGTTTGAACGCCTCGATCGTTTTCACCAAGAACTGGACACTCGTTCTAAAGCAATGGATCAAATAATGTCAATGTTCGCTATCATTCAGCCATTTTCTCTGATTTTTGCAGAAGAAGAAAAACTTCGGAAGTTTTTACCAAATATAATAGAAATTTATGATGAATTTTCTGGTGAAGATATTTTAGTGGAAATTTTTCGACTGCGGAGACATTTGAAAGCCGCTAGAATCGATCCCGAAGAAACAAAGACATGGACAGTATTGCAATTTCTGGAATTTATTGTGAAATGGGATTTTTATGAATCTCTGCCAAACTTATCCTTATGTTTAAGACTTTTCCTAACTATTTGTATATCTGTTGCTTCATGTGAAAGAAACTTTTCGAAATTAAAATTAATAAAAAGTGTTCTTCGATCAACTATGAGCGAAGATAGATTGACAAATCTGGCTATACTGTCTATTGAACATGAATATGCGAAGAAGATCAATTTTGACGAAGTCATTGACAAATTTGCAGAAGTTAAGGCTCGAAAACAGAAACTGTAATGTTATTATTCATTACTGCGACAGACCAATATGTAGGTATAAGTTTTTTCCTTTTTTCAAAAAATACATTAATGTAATTAAAAAGTATTAATCCATTACTTTTTTTCCTTTTTTGTACTGTAATATTTATTTTTTATTTTTTATACTGGCATGATTATATATACAAAGTTCAATAAAAGAAAATTTTCACTGTCTGCGTTTCTTTTCTGGCCATTATTATTATTCGTTTCATTTCATGATTATTACTGAAAATAATTTTGTCGTATAGAGGAGGGGGGTGTTAAAAAATGATCCGCTCCGGGTGTCAAATACGCTAGGTACGCCACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Zaphod3 | DOF3.6 | 2422 | 2442 | + | 16.27 | TTTATTTTTTATTTTTTATAC |
| Zaphod3 | KLF15 | 15 | 22 | - | 16.27 | CCCCGCCC |
| Zaphod3 | DOF3.2 | 2392 | 2407 | + | 16.26 | TTACTTTTTTTCCTTT |
| Zaphod3 | TSO1 | 594 | 608 | - | 16.24 | GTTTTAAATTTTAAA |
| Zaphod3 | FLC | 2399 | 2412 | - | 16.23 | ACAAAAAAGGAAAA |
| Zaphod3 | RVE6 | 1927 | 1935 | + | 16.23 | AGATATTTT |
| Zaphod3 | RVE5 | 1927 | 1935 | + | 16.21 | AGATATTTT |
| Zaphod3 | DOF5.1 | 2334 | 2352 | - | 16.18 | AAAAAGGAAAAAACTTATA |
| Zaphod3 | su(Hw) | 1616 | 1636 | + | 16.14 | GCCATTAACTATGCAACAACA |
| Zaphod3 | RVE8 | 1927 | 1935 | + | 16.08 | AGATATTTT |
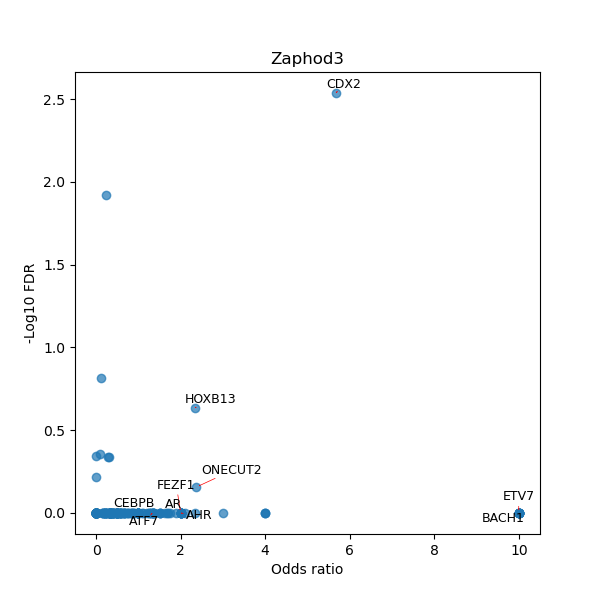
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.