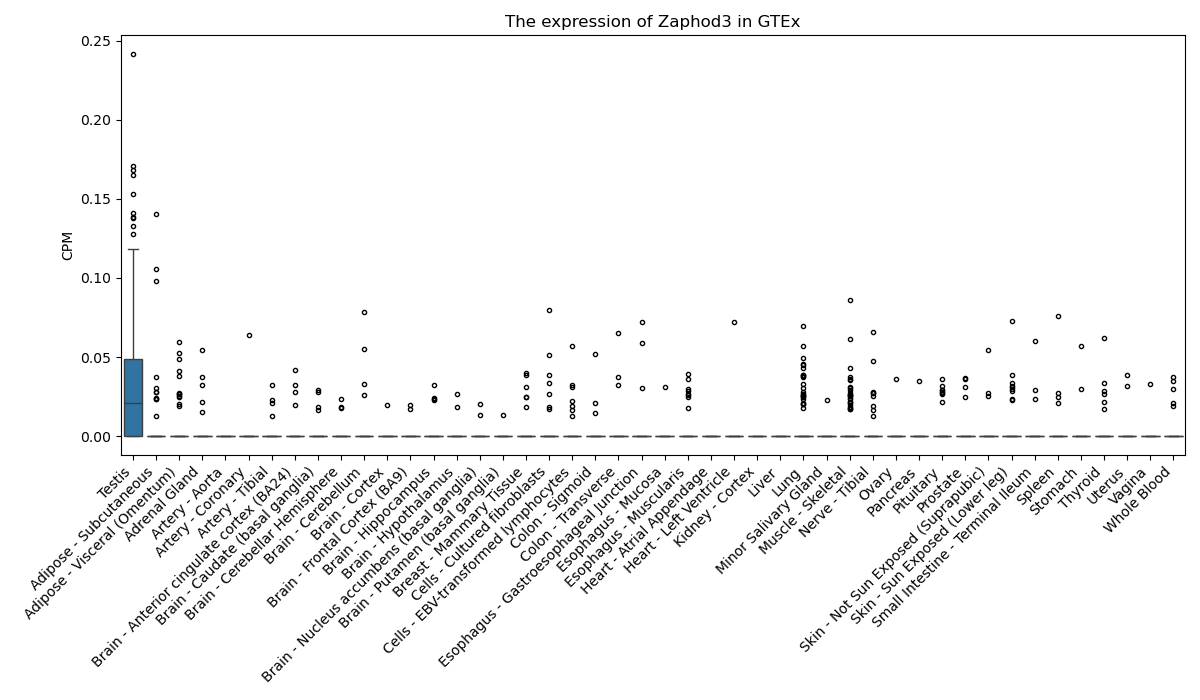
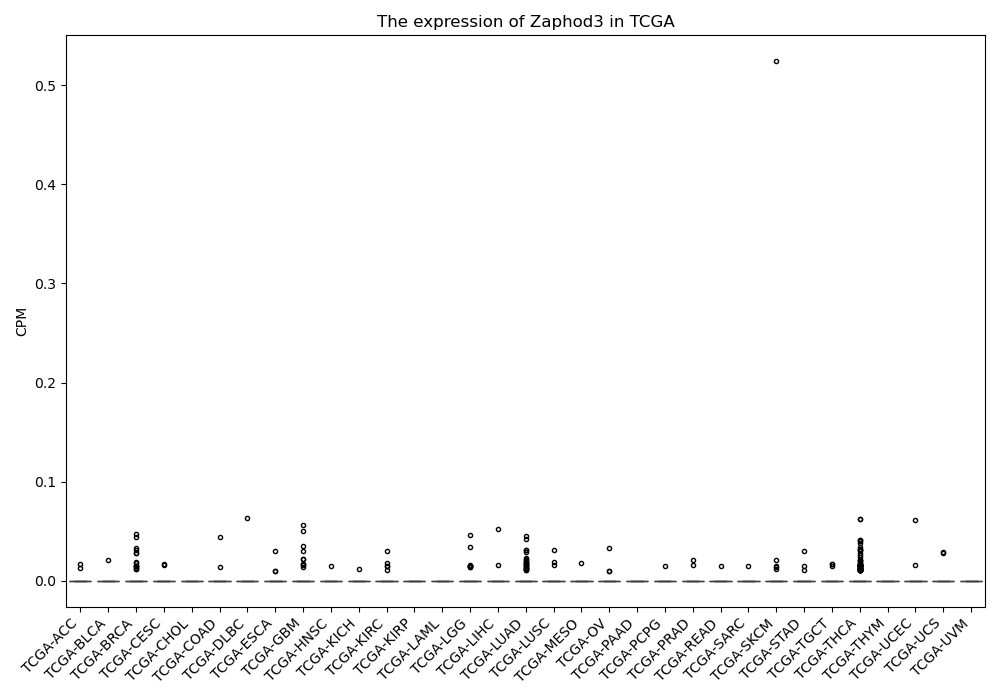
Zaphod3
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001125 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 2624 |
| Kimura value | 22.08 |
| Tau index | 1.0000 |
| Description | hAT-Tip100 DNA transposon, Zaphod3 subfamily |
| Comment | Zaphod3 encodes an ORF (~821..2293) that is roughly equally similar to that of Zaphod and Arthur. MER97a-d are non-autonomous, internal deletions of Zaphod3. |
| Sequence |
CAGTGGCGTACCAAGGGCGGGGCGGTGGGAGCGGTCCGCCCCGGGTGCAGGCAATAAGGGGGTGCATTGTCTGTAGAGAATTTAAAAACAATAATAAAACCGACTAAAAGTCGGTCTGCTTTTTATTATCACCATGCGCCGGCAATTCTAAACAATGTCAGTGATAAAATACTCCTCCCCGAAAAATCTTTTGTTGGTCTAAGTTCTAAACAATTGCTGCGGTTACTGTTGAGTTTTAATAATATATATGTAAGCTTCAAATTAGCACATTTTTATTACTTATCCTTTAATAAACATTGTATTCTACATGGAAGTTAATTCGGAGAACTCCCAGTTATACAGTCGGCCCCCGACACACGCGGACTCAGCTACACGCGTTCGTTTCGAGAGTAAGTTCGTAACGGTTCGGAATCGTTCGAGCTCGCTTCGGGCGCAGTTCGTGTCTCCAACCCCTGTGGTACTACATATTCCTGCGTTTAAACAGTAGATTCGAAATAAACAATGATAGCACAGTGATTGTAAAGACGAAGAAACAGAACTTGAGTTACTTCAATTCTGTCATTCTATGTGACCACTTGGAGTTTTTATTTGTGTTTAAAATTTAAAACAGTGAAACAGAGTGCGAACTGCGAGGTGTAATATTTTTGTTTGGTAAGTGCAAATTTTAGTTCATACATGAAATATTTTACTGAATTTGAATAATATCTTTAAAATTGAAATTTATTCTTTTTAAATTGTTAATTGTTTTAAAACTAAAGAATGAATCAAGAAAATAAAACATTACATCAGTGGTACGATTTAGTAGTGGCAGATACTGAAAGCANAATGCCTNGGAATTCTTTCTGNTAGCACTCTNGATGCTTCGAACACTGATCAAATGTGCGAAGTGGTCAGATATGNCNATATTGAAGATGGTAAAGTTGAAATAAAGGAATCATTCTTGGGCTTCTTTCCTATTTCCGGGAAGACTGNTGCTGAACTTNCAGAAGAGANNTCGAAACAACTGGACGGTGATGGGCGGGACATAANCNTCTNCTGTGGNCAAGGATATAACGATGCTGCANNTGCGGCCGGTATTCACGGTGGNGTTCANGCAAAAATCGAAGANATTAATCCCAAATCNTTATTTNTATCTTGTGCAAATCATTCTCTGAACCTTTGCGGAGTTCACTCTTTTGGAAGTATTTCTTCATGCGTGACGTTTTTTNGAACTTTGGAAAAAAATTATTCATTCTTTTCAGTCTCACCTCATCGATGGAAAACGCTGCAGAATGTAGGTATAACAGTGAAAAGACTTTCCCAGACAAGATGGAATGCTCANTATGAAGCTGTGCGCGCAGTANAGACAAATTTTGAAAAGTTAATCTCAACCTTTGAAGTACTGTGTAATCCAAAAGAAAATGTGGACNCAAGAGGATCNGCTCAGATTTTNCTCTCTGCTGTATGCGATTTTTCTTTTCTGAGTTATCTTTTTTTCTGGTGTGAAGTTTTAGATGAGGTTAATCAGACACAAAAATATTTGCAAACAGCCAGAATCAGCCTTGAACAATGTACAGTGAAACACCAAGCTTTAAAATTGTTCCTTGAAGATCGGCGCACAGAAATTGTGGAGAAGGCCATTAACTATGCAACAACAAAATGTAAGGAAATGGACATTTACATAGAAAAAAGAATCAAATTTCGAAGAAGAATGCCAGGAGAAACGACAAAAGATGCTGGTCTTACATTGCCAGAAGAAATCAAAAGGGCAATGTTTGAACGCCTCGATCGTTTTCACCAAGAACTGGACACTCGTTCTAAAGCAATGGATCAAATAATGTCAATGTTCGCTATCATTCAGCCATTTTCTCTGATTTTTGCAGAAGAAGAAAAACTTCGGAAGTTTTTACCAAATATAATAGAAATTTATGATGAATTTTCTGGTGAAGATATTTTAGTGGAAATTTTTCGACTGCGGAGACATTTGAAAGCCGCTAGAATCGATCCCGAAGAAACAAAGACATGGACAGTATTGCAATTTCTGGAATTTATTGTGAAATGGGATTTTTATGAATCTCTGCCAAACTTATCCTTATGTTTAAGACTTTTCCTAACTATTTGTATATCTGTTGCTTCATGTGAAAGAAACTTTTCGAAATTAAAATTAATAAAAAGTGTTCTTCGATCAACTATGAGCGAAGATAGATTGACAAATCTGGCTATACTGTCTATTGAACATGAATATGCGAAGAAGATCAATTTTGACGAAGTCATTGACAAATTTGCAGAAGTTAAGGCTCGAAAACAGAAACTGTAATGTTATTATTCATTACTGCGACAGACCAATATGTAGGTATAAGTTTTTTCCTTTTTTCAAAAAATACATTAATGTAATTAAAAAGTATTAATCCATTACTTTTTTTCCTTTTTTGTACTGTAATATTTATTTTTTATTTTTTATACTGGCATGATTATATATACAAAGTTCAATAAAAGAAAATTTTCACTGTCTGCGTTTCTTTTCTGGCCATTATTATTATTCGTTTCATTTCATGATTATTACTGAAAATAATTTTGTCGTATAGAGGAGGGGGGTGTTAAAAAATGATCCGCTCCGGGTGTCAAATACGCTAGGTACGCCACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Zaphod3 | Sox21b | 1813 | 1827 | - | -5.48 | AACATTGACATTATT |
| Zaphod3 | Sox21b | 209 | 223 | + | -7.44 | AACAATTGCTGCGGT |
| Zaphod3 | RARA | 1736 | 1753 | + | -7.95 | GAAATCAAAAGGGCAATG |
| Zaphod3 | Sox21b | 1813 | 1827 | + | -10.03 | AATAATGTCAATGTT |
| Zaphod3 | BPC5 | 1828 | 1857 | - | -46.74 | AAAATCAGAGAAAATGGCTGAATGATAGCG |
| Zaphod3 | BPC5 | 597 | 626 | + | -48.09 | AAAATTTAAAACAGTGAAACAGAGTGCGAA |
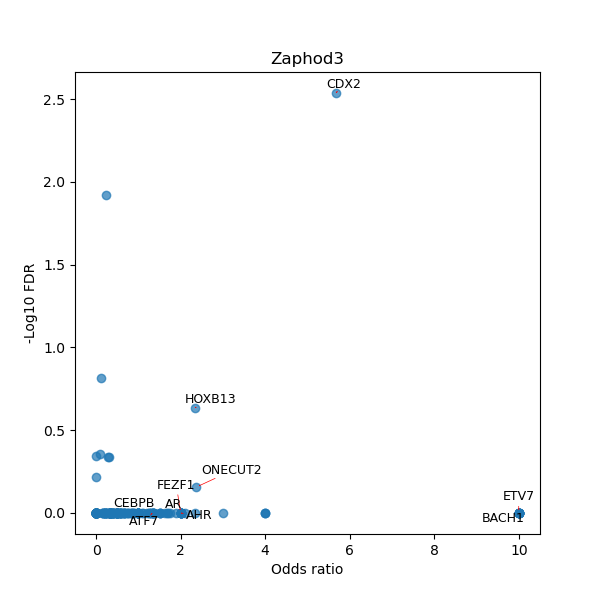
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.