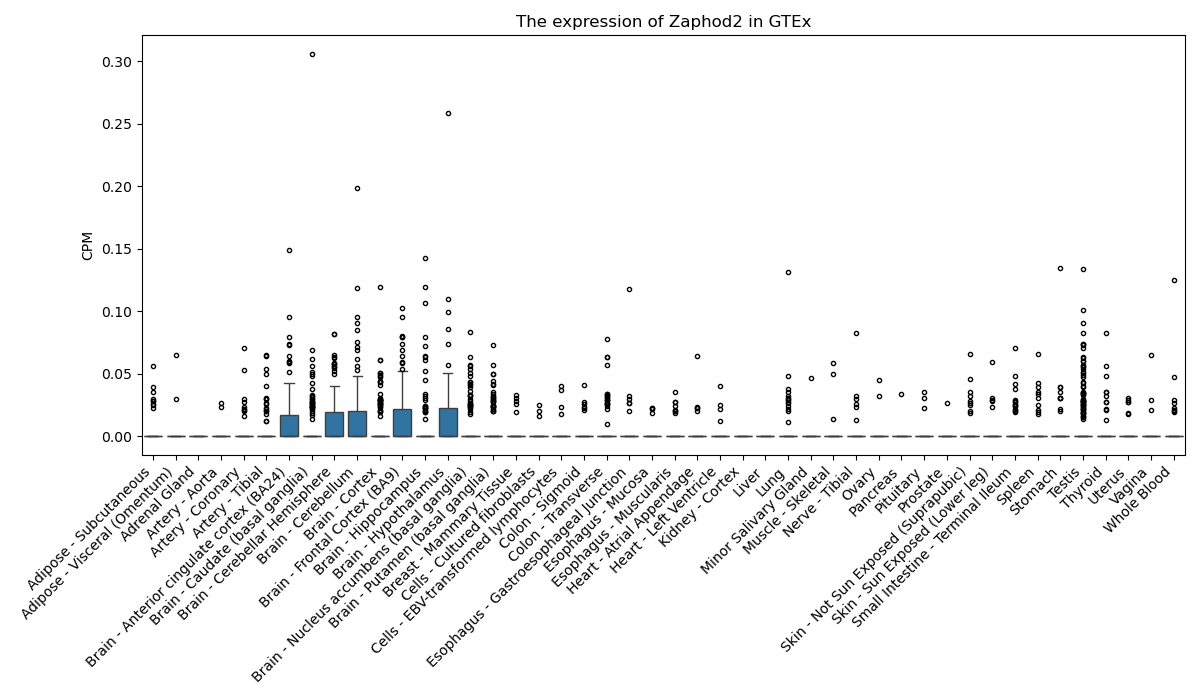
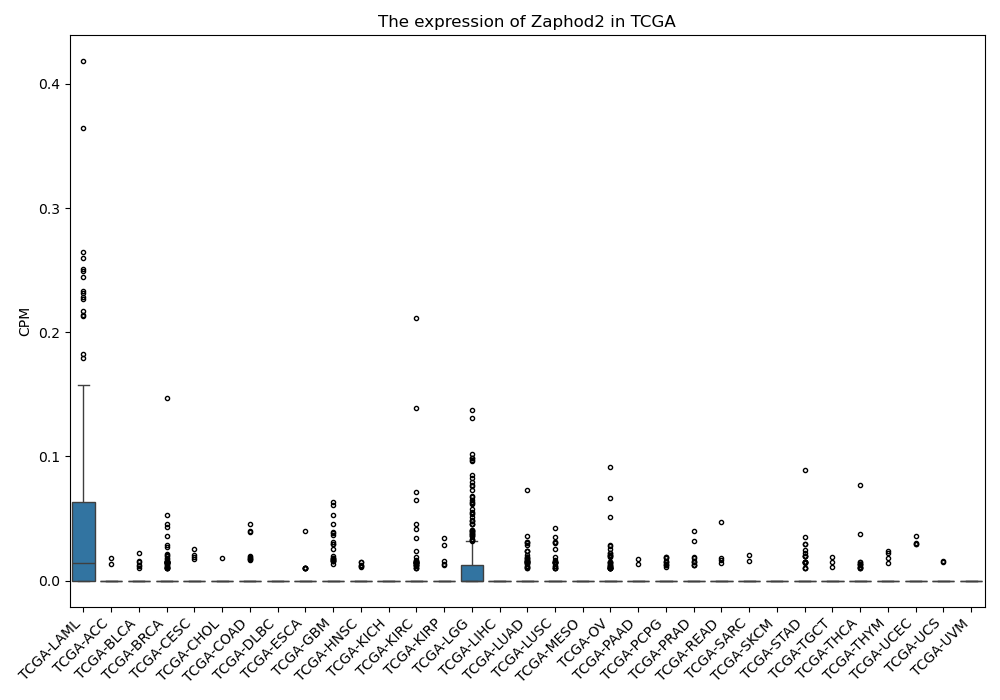
Zaphod2
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001124 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 3592 |
| Kimura value | 22.69 |
| Tau index | 0.0000 |
| Description | hAT-Tip100 DNA transposon, Zaphod2 subfamily |
| Comment | MER91A-C are short, non-autonomous derivatives of Zaphod2. |
| Sequence |
CAGGGCCGCCGCGTACGGTTGTNCAGGTTGTGCACTGCACAAGGGCGCCACATCTAAGGGGGCGCCATTCACATCGTAGACATCTTTATTATTTTGAAACATTTTTATATTTTTATTATAACTTTGTTGACTCCAAAGATGGGCGATTTGTATATTTATTATGACAATTTTCCGGCAGATGGCAGTAAAGTGTCTTGTTCTAACAAAATCAGTATATTACGACAATTTTCCGACAGATGGAAGTAAAGTGTCTTGTTCTAACGAAGTNAATATATCTCATTCCGCTTCAACTTACGAAGGAGGAACGAATTCGATAATNGNGAGACCTTGTTGGTNTAAGAATATGTAACATAACCTGTGCTTCTNAACAAGGAATTGCTTTTCCGACTTCTGCACTNNTCAGTAGGTATCTTNAAATNNNAANAAANAATCTCCTATTGGTATTGGTGCACTGGCTAAGTTTGACGTTTCGTATTGCAGTTCNTATNATCTGCCATTTGTTTACGGTACCAGAAAGGTATTGCTGAGTTCCCGGTTTAAANCTACAGNTACTTTCNTTATGACACGAATCACTATGTAATAACTAATATTCATTTACCATTAATAATCCACAAAGTAAGTATTACATAGACATAAATTAGAGATTACTATGTAATGATCCGCTAATTATATTATAGCAATTGTTTATTTGTTGTTATGGAAATTTCGTCAGTACAAAAATTAACTGCTGTTAAAATTTAGCGGCTTNTTACATACATATTATTTTAAAATGTTACAAATTCTAAGATAAAATACTAAGCTATTATATTATTTCTCCCCAGATGTCGGAAAAAAATTTTAAAAAATTATCAGNATCAGAAAACAGAAAATGGAAAGCCATTAGAGAAGAAAATGAAAAAAAATTATTTGGATCATTGAATAAATTTTTGGAGAAAATAGATGAAATGCCAGAAAAAAAGGAAAAGACTTCAGAGCTACTAGTTTCAGCAGAATGTAGCACAGATACGTCACTGATGGAGCAGGATGTGAATTTACCTTCTACTTCCATGTCAACCATAACTGATGAATTTCAAGAAAAACTGGAAGAGAAAAAATCAGAAGAATCCATTAAACCGAATGTACTTTCTGCCGATCCAGGTACGTGGCCATGTAATTTGAATATTAAACAACGGNACACTCTTGTTGAAAATTATCCACCTCAAGTAAGAAATTTTAACTTTCCAAAAGACAGTACAGGTAGGAAGTTTTCGGAAACTTATTANACACGAATTCTTCCAAATGGTGAAAAAATCACTAGATCCTGGTTACTTTATTCTACCTCAAAGGATTCTGTATTTTGTCTATATTGCAAACTCTTTGGGGATAGAAAAAATCAACTGAAGAATGAGAATGGATGCAAAGATTGGCAGCATTTATCACATATTCTTAGTAAACATGAAGAAAGTGAAATGCACATCAACAATAGTGTTAAGTATTCAAAATTGAAATCTGATTTGAAAAAATATATAACTATCGATGCTGCAGAACAGAGACTATATGAAAATGAGGAAAAATATTGGCGTGCTGTTGTTGAACGCATAATATACATGATTCAGTTTTTGGCAAGGCAATGTCTAGCGTTCAGAGGATCATCCAAGAAANTTTATGAGCGTGATAATGGGAATTTTCTTCATTTGGCAGAAACCGTAGCAAAATTTGATAATATTATGGCAGAACATTTATGCAGAATACAAACATTGACACCAAATAATATTACACATTATTTAGGGGATGAAATACAAAACCAAATAATTAAAATTATAGCAGATGCGATGAATGAAATCATTTTGAATAAGGTAAGAAATNCTAAATATTACTCAATTATTTTAGATACAACCCCTGATGTAAGCCATGTAGAACAGATAACAATTATTATTAGATTTGTGGATTTCCAAAATGCAAATATTTCTATATGTAAACATTTTGGGGGATTTTGTCCAGTAGATGACACAACTGGAGAAGGATTATATACATTTTTAGTAGACTACTTACAAAAACCGAATCTAGACAAAAATAATTTGAGAGGCCAGGGATACGACAATGCCACAAATATGCATGGAAAACATAACGGATTGCAACAGAAGATCCTCGAAAAATATTCTAGGGCATTCTATATTCCTTGTTCTGCGCATTCTTTAAACTTGGTAGTGAACGATGCAGCAAAAATCTCTTTTGACACCACTGAATTTTTTTATATNATCCAAGAACTATACAACTACTTTTCAGCTTCAACGAAACGTTGGAGTGTATTGAAAACACATTTACCAAACATAAATTTGAAACCATTGTCGAATACGCGATGGGAAAGTCGAGTTGATGCCATCATGCCACTAAAAACTCAATTAGAGGCAGTATGCAATGCTCTTTTAGAAATTNTTGATGATATTAATAAAGATATAGATTCTAGAATACAAGCCAAAGNTATACTTTCGAAAATATCCACTTTTAAATTTATTTGCTCAGTACTAATTTGGCATGATGTTTTAAGTAAAATTAATGTTATAAGCAAAATGATGCAGGAACCAAAATTCAATGTACATAATGCCCTTGAAAGCATCAAAGGGATTAAGAAATTCTTTGAAGAAAAACGAAGCGATGATGTTTATGAAAAATATATTATTGATGCTAAACAAATTGCAGAGTCTCTTGACATTGACGCCAACTTCTCCAATGTAGAAAATACTAGAAAAAGGAGAAAGAAGACTCATTTCGACTGCAAATCCCACGATCAGCCCATCTATGATCCAAAATTAAATTTCAAAATCAATTTCTTTTTTTCTATATTGGATACAACAATATCATCTTTAAAAGANAGGTTTACGATACCGAAAGAATATCATGGCCTTTTCAGTGTTCTAGATGACATGTGTAATTTGAAGAATTATAATGAATATGAACTACTGCAACTATGTAAAAATTTACAATTGAAATTGACTGATGAACATAGTAAGGAATGTGATATTGATGGGACATCAACGTACGAGGAACTAAAANCCTTAAGTTGTTTTCTAAAATCTAGTATNACTCCCGTTGAAGTATTGCAATATATTTGCAGCAATAATTTACCGGACACATTTCCGAATACTTTTGTAGCTCTCAGGATATATTTAACTTTGCCAATATCGGTCGCCAGTGGAGAACGTAGCTTCTCAAAATTAAAATTTATTAAAAACTACTTAAGGTCTACAATGTCACAAGAAAGACTGTCAAATCTAGCGACCATTTCTATAGAAAACGAGATATTGGAGCAAGTAAGCATTGAAACTGTCATAAAGAATTTTATTTCCATGAAAGCAAGACGTGTAACTATTTTATAACAAATTATTATATCCTAGTGACTTTGAGTTCCTAATTATTGTGATTAAAANTTCGAATTTTCTATTATTTTATGTTCAATAATTTGATTTGTATAATTTATACAGAATGACTTTATATTTGTTGTAATTGTTATTATTGAGAAATAAATAATATACTAACTTNAAAATTTTACTTGAGGAAGGGGCGCCTTTTTCTAATTCGCACAAAGGCGCCGTATGGGCTAGCAGCGGCCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Zaphod2 | dl | 1923 | 1932 | + | 15.17 | GTGGATTTCC |
| Zaphod2 | AP3 | 1665 | 1677 | - | 15.17 | CCAAATGAAGAAA |
| Zaphod2 | SMZ | 3017 | 3024 | - | 15.13 | CCTCGTAC |
| Zaphod2 | SFP1 | 830 | 839 | + | 15.11 | AAAAAATTTT |
| Zaphod2 | SOL1 | 2788 | 2802 | + | 15.10 | AATTAAATTTCAAAA |
| Zaphod2 | CTCF | 171 | 185 | + | 15.10 | TCCGGCAGATGGCAG |
| Zaphod2 | CUP2 | 949 | 959 | + | 15.09 | CAGAAAAAAAG |
| Zaphod2 | HSFA1E | 2714 | 2727 | + | 15.08 | AGAAAATACTAGAA |
| Zaphod2 | Foxj3 | 497 | 505 | - | 15.06 | GTAAACAAA |
| Zaphod2 | FKH1 | 497 | 509 | - | 14.92 | TACCGTAAACAAA |
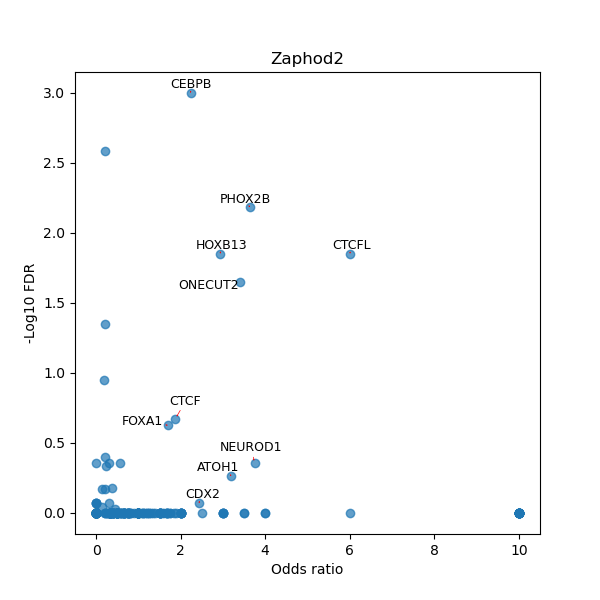
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.