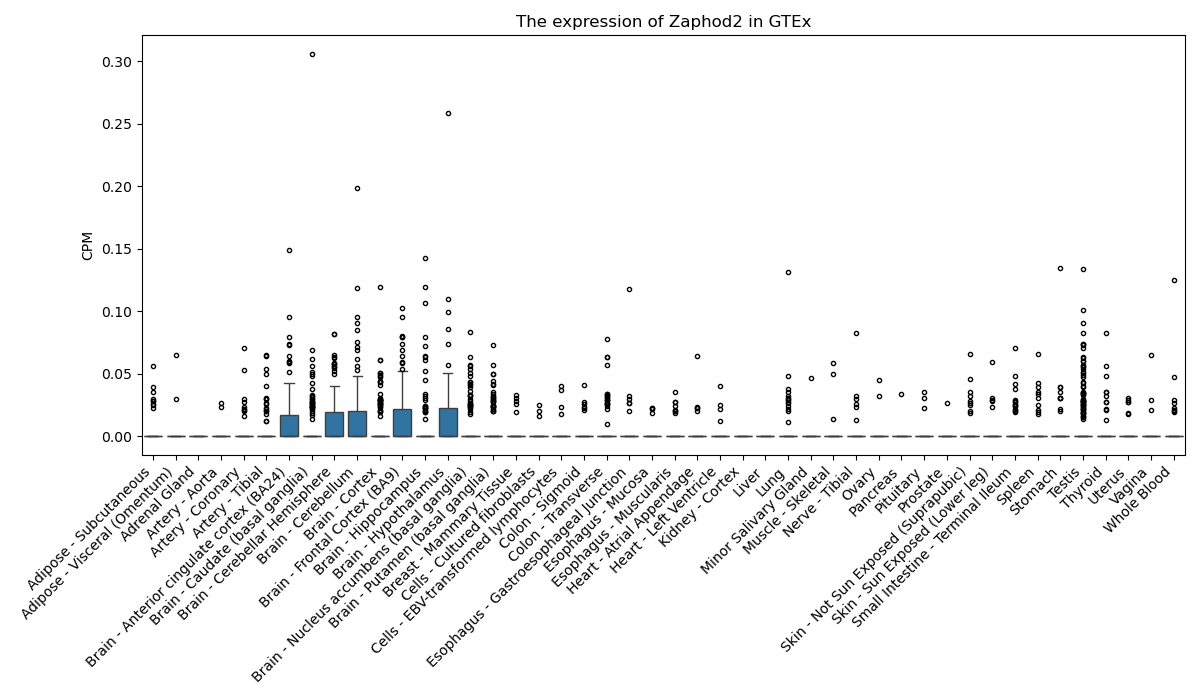
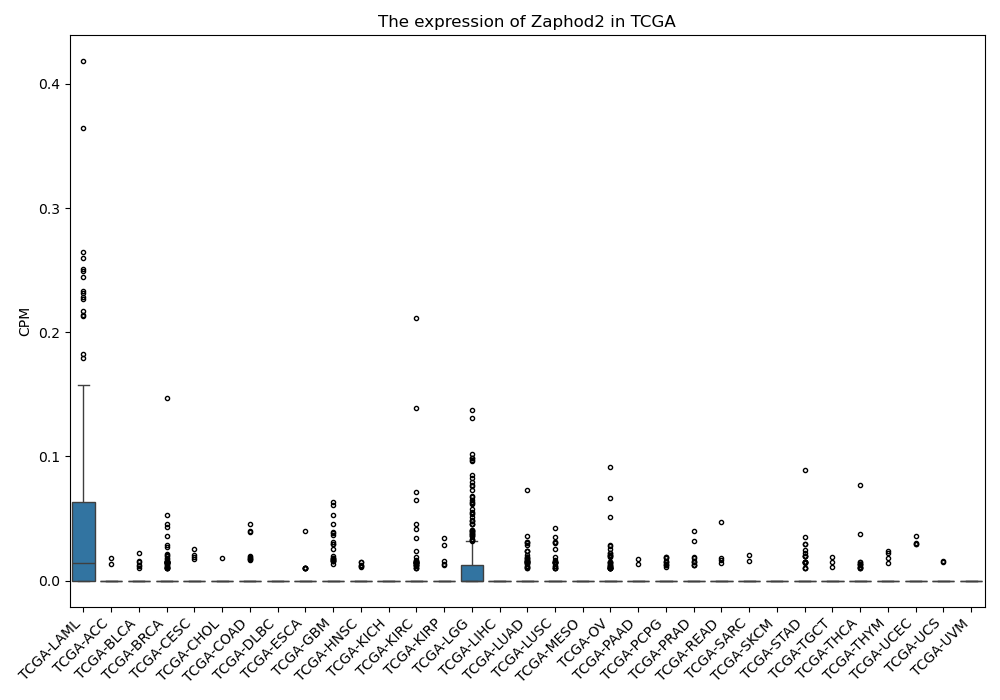
Zaphod2
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001124 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 3592 |
| Kimura value | 22.69 |
| Tau index | 0.0000 |
| Description | hAT-Tip100 DNA transposon, Zaphod2 subfamily |
| Comment | MER91A-C are short, non-autonomous derivatives of Zaphod2. |
| Sequence |
CAGGGCCGCCGCGTACGGTTGTNCAGGTTGTGCACTGCACAAGGGCGCCACATCTAAGGGGGCGCCATTCACATCGTAGACATCTTTATTATTTTGAAACATTTTTATATTTTTATTATAACTTTGTTGACTCCAAAGATGGGCGATTTGTATATTTATTATGACAATTTTCCGGCAGATGGCAGTAAAGTGTCTTGTTCTAACAAAATCAGTATATTACGACAATTTTCCGACAGATGGAAGTAAAGTGTCTTGTTCTAACGAAGTNAATATATCTCATTCCGCTTCAACTTACGAAGGAGGAACGAATTCGATAATNGNGAGACCTTGTTGGTNTAAGAATATGTAACATAACCTGTGCTTCTNAACAAGGAATTGCTTTTCCGACTTCTGCACTNNTCAGTAGGTATCTTNAAATNNNAANAAANAATCTCCTATTGGTATTGGTGCACTGGCTAAGTTTGACGTTTCGTATTGCAGTTCNTATNATCTGCCATTTGTTTACGGTACCAGAAAGGTATTGCTGAGTTCCCGGTTTAAANCTACAGNTACTTTCNTTATGACACGAATCACTATGTAATAACTAATATTCATTTACCATTAATAATCCACAAAGTAAGTATTACATAGACATAAATTAGAGATTACTATGTAATGATCCGCTAATTATATTATAGCAATTGTTTATTTGTTGTTATGGAAATTTCGTCAGTACAAAAATTAACTGCTGTTAAAATTTAGCGGCTTNTTACATACATATTATTTTAAAATGTTACAAATTCTAAGATAAAATACTAAGCTATTATATTATTTCTCCCCAGATGTCGGAAAAAAATTTTAAAAAATTATCAGNATCAGAAAACAGAAAATGGAAAGCCATTAGAGAAGAAAATGAAAAAAAATTATTTGGATCATTGAATAAATTTTTGGAGAAAATAGATGAAATGCCAGAAAAAAAGGAAAAGACTTCAGAGCTACTAGTTTCAGCAGAATGTAGCACAGATACGTCACTGATGGAGCAGGATGTGAATTTACCTTCTACTTCCATGTCAACCATAACTGATGAATTTCAAGAAAAACTGGAAGAGAAAAAATCAGAAGAATCCATTAAACCGAATGTACTTTCTGCCGATCCAGGTACGTGGCCATGTAATTTGAATATTAAACAACGGNACACTCTTGTTGAAAATTATCCACCTCAAGTAAGAAATTTTAACTTTCCAAAAGACAGTACAGGTAGGAAGTTTTCGGAAACTTATTANACACGAATTCTTCCAAATGGTGAAAAAATCACTAGATCCTGGTTACTTTATTCTACCTCAAAGGATTCTGTATTTTGTCTATATTGCAAACTCTTTGGGGATAGAAAAAATCAACTGAAGAATGAGAATGGATGCAAAGATTGGCAGCATTTATCACATATTCTTAGTAAACATGAAGAAAGTGAAATGCACATCAACAATAGTGTTAAGTATTCAAAATTGAAATCTGATTTGAAAAAATATATAACTATCGATGCTGCAGAACAGAGACTATATGAAAATGAGGAAAAATATTGGCGTGCTGTTGTTGAACGCATAATATACATGATTCAGTTTTTGGCAAGGCAATGTCTAGCGTTCAGAGGATCATCCAAGAAANTTTATGAGCGTGATAATGGGAATTTTCTTCATTTGGCAGAAACCGTAGCAAAATTTGATAATATTATGGCAGAACATTTATGCAGAATACAAACATTGACACCAAATAATATTACACATTATTTAGGGGATGAAATACAAAACCAAATAATTAAAATTATAGCAGATGCGATGAATGAAATCATTTTGAATAAGGTAAGAAATNCTAAATATTACTCAATTATTTTAGATACAACCCCTGATGTAAGCCATGTAGAACAGATAACAATTATTATTAGATTTGTGGATTTCCAAAATGCAAATATTTCTATATGTAAACATTTTGGGGGATTTTGTCCAGTAGATGACACAACTGGAGAAGGATTATATACATTTTTAGTAGACTACTTACAAAAACCGAATCTAGACAAAAATAATTTGAGAGGCCAGGGATACGACAATGCCACAAATATGCATGGAAAACATAACGGATTGCAACAGAAGATCCTCGAAAAATATTCTAGGGCATTCTATATTCCTTGTTCTGCGCATTCTTTAAACTTGGTAGTGAACGATGCAGCAAAAATCTCTTTTGACACCACTGAATTTTTTTATATNATCCAAGAACTATACAACTACTTTTCAGCTTCAACGAAACGTTGGAGTGTATTGAAAACACATTTACCAAACATAAATTTGAAACCATTGTCGAATACGCGATGGGAAAGTCGAGTTGATGCCATCATGCCACTAAAAACTCAATTAGAGGCAGTATGCAATGCTCTTTTAGAAATTNTTGATGATATTAATAAAGATATAGATTCTAGAATACAAGCCAAAGNTATACTTTCGAAAATATCCACTTTTAAATTTATTTGCTCAGTACTAATTTGGCATGATGTTTTAAGTAAAATTAATGTTATAAGCAAAATGATGCAGGAACCAAAATTCAATGTACATAATGCCCTTGAAAGCATCAAAGGGATTAAGAAATTCTTTGAAGAAAAACGAAGCGATGATGTTTATGAAAAATATATTATTGATGCTAAACAAATTGCAGAGTCTCTTGACATTGACGCCAACTTCTCCAATGTAGAAAATACTAGAAAAAGGAGAAAGAAGACTCATTTCGACTGCAAATCCCACGATCAGCCCATCTATGATCCAAAATTAAATTTCAAAATCAATTTCTTTTTTTCTATATTGGATACAACAATATCATCTTTAAAAGANAGGTTTACGATACCGAAAGAATATCATGGCCTTTTCAGTGTTCTAGATGACATGTGTAATTTGAAGAATTATAATGAATATGAACTACTGCAACTATGTAAAAATTTACAATTGAAATTGACTGATGAACATAGTAAGGAATGTGATATTGATGGGACATCAACGTACGAGGAACTAAAANCCTTAAGTTGTTTTCTAAAATCTAGTATNACTCCCGTTGAAGTATTGCAATATATTTGCAGCAATAATTTACCGGACACATTTCCGAATACTTTTGTAGCTCTCAGGATATATTTAACTTTGCCAATATCGGTCGCCAGTGGAGAACGTAGCTTCTCAAAATTAAAATTTATTAAAAACTACTTAAGGTCTACAATGTCACAAGAAAGACTGTCAAATCTAGCGACCATTTCTATAGAAAACGAGATATTGGAGCAAGTAAGCATTGAAACTGTCATAAAGAATTTTATTTCCATGAAAGCAAGACGTGTAACTATTTTATAACAAATTATTATATCCTAGTGACTTTGAGTTCCTAATTATTGTGATTAAAANTTCGAATTTTCTATTATTTTATGTTCAATAATTTGATTTGTATAATTTATACAGAATGACTTTATATTTGTTGTAATTGTTATTATTGAGAAATAAATAATATACTAACTTNAAAATTTTACTTGAGGAAGGGGCGCCTTTTTCTAATTCGCACAAAGGCGCCGTATGGGCTAGCAGCGGCCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Zaphod2 | TCX3 | 1151 | 1162 | - | 15.43 | ATATTCAAATTA |
| Zaphod2 | DOF4.2 | 954 | 964 | + | 15.42 | AAAAAGGAAAA |
| Zaphod2 | SIX2 | 1483 | 1493 | + | 15.35 | TGAAATCTGAT |
| Zaphod2 | Zm00001d002364 | 4 | 13 | + | 15.33 | GGCCGCCGCG |
| Zaphod2 | E2F2 | 41 | 53 | - | 15.29 | ATGTGGCGCCCTT |
| Zaphod2 | Msgn1 | 493 | 502 | - | 15.29 | AACAAATGGC |
| Zaphod2 | FOXF2 | 498 | 506 | - | 15.28 | CGTAAACAA |
| Zaphod2 | TSO1 | 2787 | 2801 | + | 15.28 | AAATTAAATTTCAAA |
| Zaphod2 | CHES-1-like | 497 | 506 | - | 15.27 | CGTAAACAAA |
| Zaphod2 | FOXN3 | 498 | 505 | - | 15.19 | GTAAACAA |
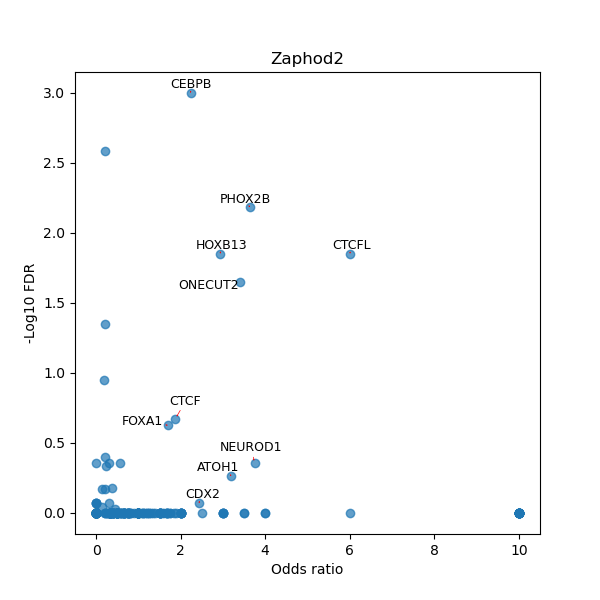
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.