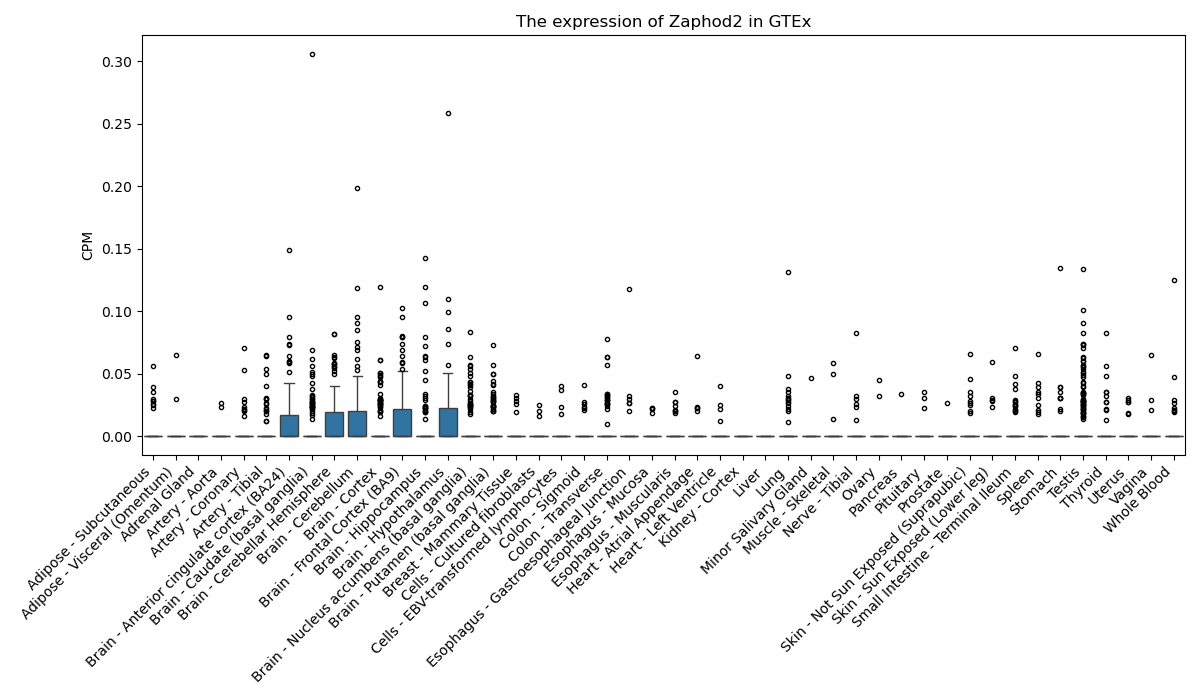
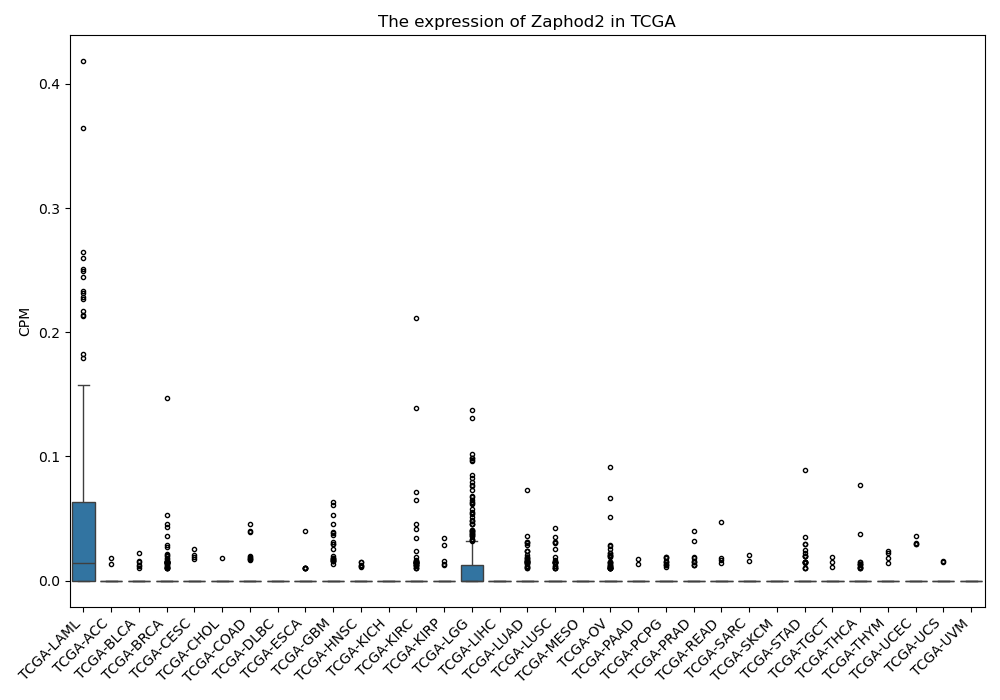
Zaphod2
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001124 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 3592 |
| Kimura value | 22.69 |
| Tau index | 0.0000 |
| Description | hAT-Tip100 DNA transposon, Zaphod2 subfamily |
| Comment | MER91A-C are short, non-autonomous derivatives of Zaphod2. |
| Sequence |
CAGGGCCGCCGCGTACGGTTGTNCAGGTTGTGCACTGCACAAGGGCGCCACATCTAAGGGGGCGCCATTCACATCGTAGACATCTTTATTATTTTGAAACATTTTTATATTTTTATTATAACTTTGTTGACTCCAAAGATGGGCGATTTGTATATTTATTATGACAATTTTCCGGCAGATGGCAGTAAAGTGTCTTGTTCTAACAAAATCAGTATATTACGACAATTTTCCGACAGATGGAAGTAAAGTGTCTTGTTCTAACGAAGTNAATATATCTCATTCCGCTTCAACTTACGAAGGAGGAACGAATTCGATAATNGNGAGACCTTGTTGGTNTAAGAATATGTAACATAACCTGTGCTTCTNAACAAGGAATTGCTTTTCCGACTTCTGCACTNNTCAGTAGGTATCTTNAAATNNNAANAAANAATCTCCTATTGGTATTGGTGCACTGGCTAAGTTTGACGTTTCGTATTGCAGTTCNTATNATCTGCCATTTGTTTACGGTACCAGAAAGGTATTGCTGAGTTCCCGGTTTAAANCTACAGNTACTTTCNTTATGACACGAATCACTATGTAATAACTAATATTCATTTACCATTAATAATCCACAAAGTAAGTATTACATAGACATAAATTAGAGATTACTATGTAATGATCCGCTAATTATATTATAGCAATTGTTTATTTGTTGTTATGGAAATTTCGTCAGTACAAAAATTAACTGCTGTTAAAATTTAGCGGCTTNTTACATACATATTATTTTAAAATGTTACAAATTCTAAGATAAAATACTAAGCTATTATATTATTTCTCCCCAGATGTCGGAAAAAAATTTTAAAAAATTATCAGNATCAGAAAACAGAAAATGGAAAGCCATTAGAGAAGAAAATGAAAAAAAATTATTTGGATCATTGAATAAATTTTTGGAGAAAATAGATGAAATGCCAGAAAAAAAGGAAAAGACTTCAGAGCTACTAGTTTCAGCAGAATGTAGCACAGATACGTCACTGATGGAGCAGGATGTGAATTTACCTTCTACTTCCATGTCAACCATAACTGATGAATTTCAAGAAAAACTGGAAGAGAAAAAATCAGAAGAATCCATTAAACCGAATGTACTTTCTGCCGATCCAGGTACGTGGCCATGTAATTTGAATATTAAACAACGGNACACTCTTGTTGAAAATTATCCACCTCAAGTAAGAAATTTTAACTTTCCAAAAGACAGTACAGGTAGGAAGTTTTCGGAAACTTATTANACACGAATTCTTCCAAATGGTGAAAAAATCACTAGATCCTGGTTACTTTATTCTACCTCAAAGGATTCTGTATTTTGTCTATATTGCAAACTCTTTGGGGATAGAAAAAATCAACTGAAGAATGAGAATGGATGCAAAGATTGGCAGCATTTATCACATATTCTTAGTAAACATGAAGAAAGTGAAATGCACATCAACAATAGTGTTAAGTATTCAAAATTGAAATCTGATTTGAAAAAATATATAACTATCGATGCTGCAGAACAGAGACTATATGAAAATGAGGAAAAATATTGGCGTGCTGTTGTTGAACGCATAATATACATGATTCAGTTTTTGGCAAGGCAATGTCTAGCGTTCAGAGGATCATCCAAGAAANTTTATGAGCGTGATAATGGGAATTTTCTTCATTTGGCAGAAACCGTAGCAAAATTTGATAATATTATGGCAGAACATTTATGCAGAATACAAACATTGACACCAAATAATATTACACATTATTTAGGGGATGAAATACAAAACCAAATAATTAAAATTATAGCAGATGCGATGAATGAAATCATTTTGAATAAGGTAAGAAATNCTAAATATTACTCAATTATTTTAGATACAACCCCTGATGTAAGCCATGTAGAACAGATAACAATTATTATTAGATTTGTGGATTTCCAAAATGCAAATATTTCTATATGTAAACATTTTGGGGGATTTTGTCCAGTAGATGACACAACTGGAGAAGGATTATATACATTTTTAGTAGACTACTTACAAAAACCGAATCTAGACAAAAATAATTTGAGAGGCCAGGGATACGACAATGCCACAAATATGCATGGAAAACATAACGGATTGCAACAGAAGATCCTCGAAAAATATTCTAGGGCATTCTATATTCCTTGTTCTGCGCATTCTTTAAACTTGGTAGTGAACGATGCAGCAAAAATCTCTTTTGACACCACTGAATTTTTTTATATNATCCAAGAACTATACAACTACTTTTCAGCTTCAACGAAACGTTGGAGTGTATTGAAAACACATTTACCAAACATAAATTTGAAACCATTGTCGAATACGCGATGGGAAAGTCGAGTTGATGCCATCATGCCACTAAAAACTCAATTAGAGGCAGTATGCAATGCTCTTTTAGAAATTNTTGATGATATTAATAAAGATATAGATTCTAGAATACAAGCCAAAGNTATACTTTCGAAAATATCCACTTTTAAATTTATTTGCTCAGTACTAATTTGGCATGATGTTTTAAGTAAAATTAATGTTATAAGCAAAATGATGCAGGAACCAAAATTCAATGTACATAATGCCCTTGAAAGCATCAAAGGGATTAAGAAATTCTTTGAAGAAAAACGAAGCGATGATGTTTATGAAAAATATATTATTGATGCTAAACAAATTGCAGAGTCTCTTGACATTGACGCCAACTTCTCCAATGTAGAAAATACTAGAAAAAGGAGAAAGAAGACTCATTTCGACTGCAAATCCCACGATCAGCCCATCTATGATCCAAAATTAAATTTCAAAATCAATTTCTTTTTTTCTATATTGGATACAACAATATCATCTTTAAAAGANAGGTTTACGATACCGAAAGAATATCATGGCCTTTTCAGTGTTCTAGATGACATGTGTAATTTGAAGAATTATAATGAATATGAACTACTGCAACTATGTAAAAATTTACAATTGAAATTGACTGATGAACATAGTAAGGAATGTGATATTGATGGGACATCAACGTACGAGGAACTAAAANCCTTAAGTTGTTTTCTAAAATCTAGTATNACTCCCGTTGAAGTATTGCAATATATTTGCAGCAATAATTTACCGGACACATTTCCGAATACTTTTGTAGCTCTCAGGATATATTTAACTTTGCCAATATCGGTCGCCAGTGGAGAACGTAGCTTCTCAAAATTAAAATTTATTAAAAACTACTTAAGGTCTACAATGTCACAAGAAAGACTGTCAAATCTAGCGACCATTTCTATAGAAAACGAGATATTGGAGCAAGTAAGCATTGAAACTGTCATAAAGAATTTTATTTCCATGAAAGCAAGACGTGTAACTATTTTATAACAAATTATTATATCCTAGTGACTTTGAGTTCCTAATTATTGTGATTAAAANTTCGAATTTTCTATTATTTTATGTTCAATAATTTGATTTGTATAATTTATACAGAATGACTTTATATTTGTTGTAATTGTTATTATTGAGAAATAAATAATATACTAACTTNAAAATTTTACTTGAGGAAGGGGCGCCTTTTTCTAATTCGCACAAAGGCGCCGTATGGGCTAGCAGCGGCCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Zaphod2 | Foxq1 | 680 | 689 | + | 15.87 | ATTGTTTATT |
| Zaphod2 | Foxl2 | 1951 | 1960 | + | 15.83 | TATGTAAACA |
| Zaphod2 | FOXB1 | 1951 | 1961 | + | 15.71 | TATGTAAACAT |
| Zaphod2 | Stat2 | 859 | 868 | + | 15.65 | AAAACAGAAA |
| Zaphod2 | TCX2 | 1153 | 1167 | - | 15.58 | GTTTAATATTCAAAT |
| Zaphod2 | DOF1.5 | 949 | 964 | + | 15.56 | CAGAAAAAAAGGAAAA |
| Zaphod2 | Zm00001d052229 | 4 | 13 | + | 15.54 | GGCCGCCGCG |
| Zaphod2 | Irf1 | 1439 | 1449 | + | 15.53 | AGAAAGTGAAA |
| Zaphod2 | STB3 | 1281 | 1291 | - | 15.48 | ATTTTTTCACC |
| Zaphod2 | FOXC1 | 1951 | 1961 | + | 15.44 | TATGTAAACAT |
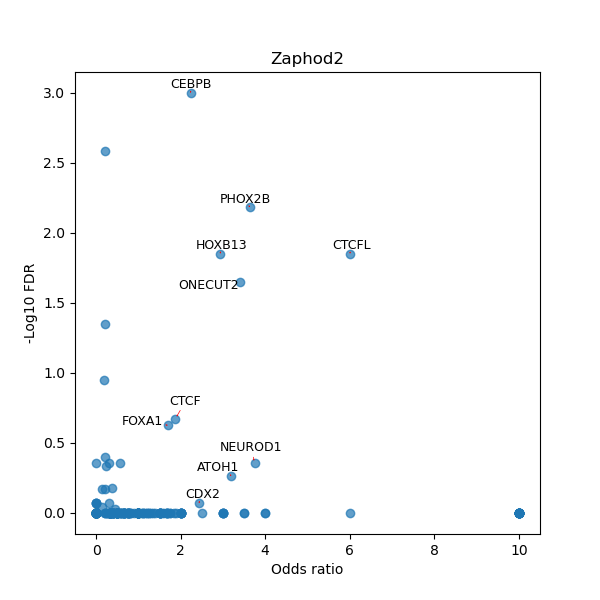
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.