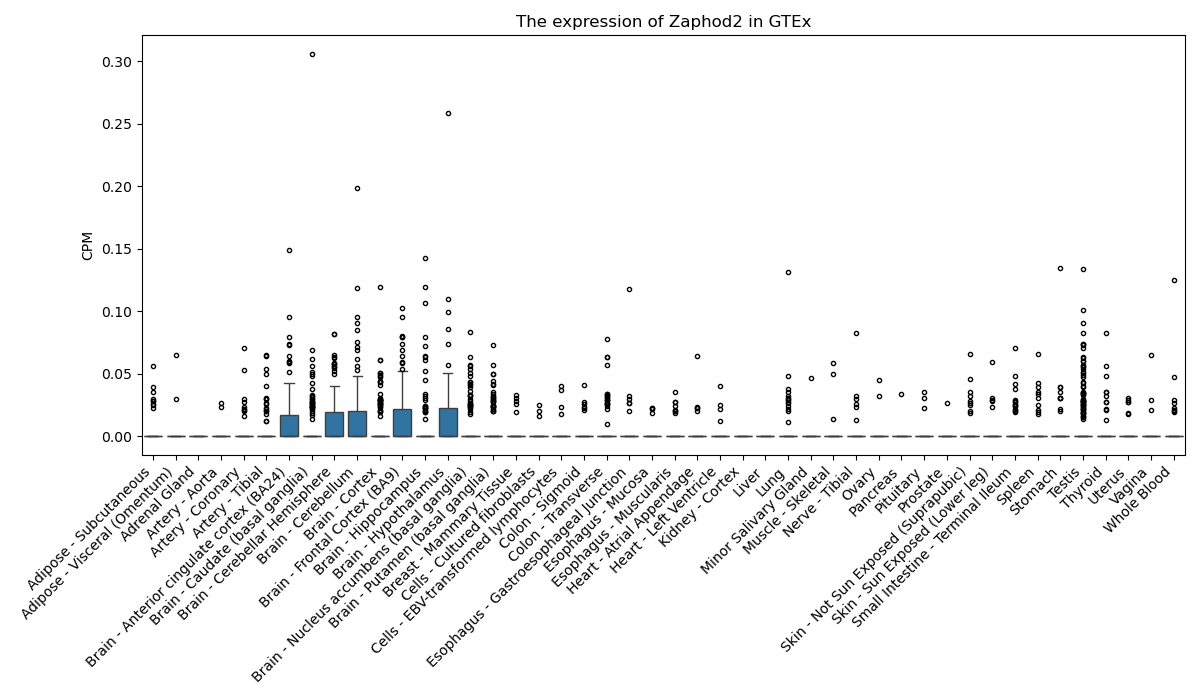
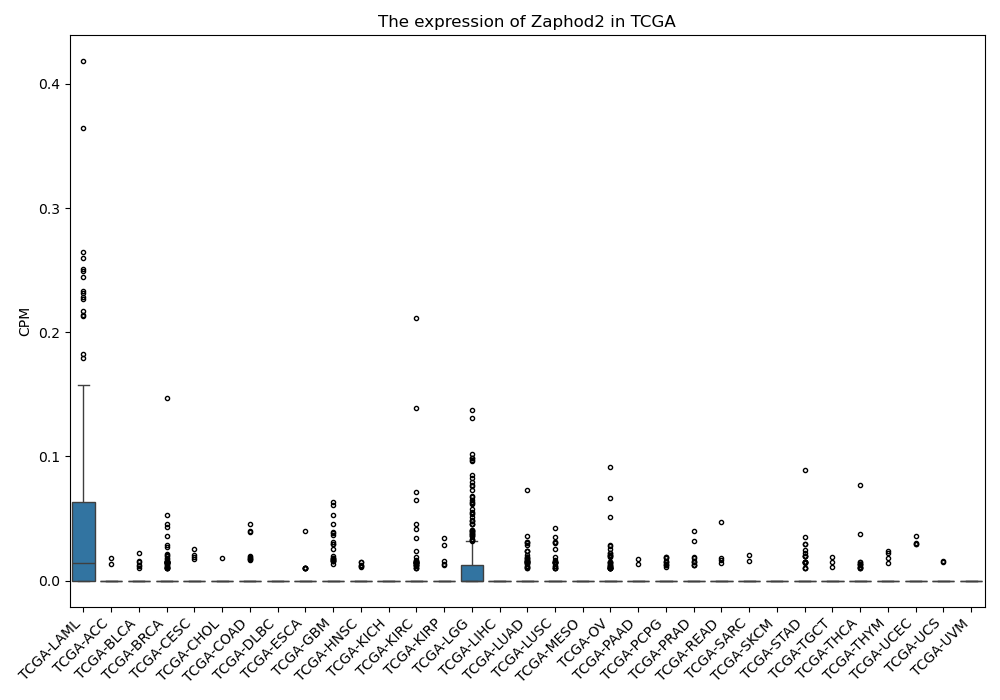
Zaphod2
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001124 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 3592 |
| Kimura value | 22.69 |
| Tau index | 0.0000 |
| Description | hAT-Tip100 DNA transposon, Zaphod2 subfamily |
| Comment | MER91A-C are short, non-autonomous derivatives of Zaphod2. |
| Sequence |
CAGGGCCGCCGCGTACGGTTGTNCAGGTTGTGCACTGCACAAGGGCGCCACATCTAAGGGGGCGCCATTCACATCGTAGACATCTTTATTATTTTGAAACATTTTTATATTTTTATTATAACTTTGTTGACTCCAAAGATGGGCGATTTGTATATTTATTATGACAATTTTCCGGCAGATGGCAGTAAAGTGTCTTGTTCTAACAAAATCAGTATATTACGACAATTTTCCGACAGATGGAAGTAAAGTGTCTTGTTCTAACGAAGTNAATATATCTCATTCCGCTTCAACTTACGAAGGAGGAACGAATTCGATAATNGNGAGACCTTGTTGGTNTAAGAATATGTAACATAACCTGTGCTTCTNAACAAGGAATTGCTTTTCCGACTTCTGCACTNNTCAGTAGGTATCTTNAAATNNNAANAAANAATCTCCTATTGGTATTGGTGCACTGGCTAAGTTTGACGTTTCGTATTGCAGTTCNTATNATCTGCCATTTGTTTACGGTACCAGAAAGGTATTGCTGAGTTCCCGGTTTAAANCTACAGNTACTTTCNTTATGACACGAATCACTATGTAATAACTAATATTCATTTACCATTAATAATCCACAAAGTAAGTATTACATAGACATAAATTAGAGATTACTATGTAATGATCCGCTAATTATATTATAGCAATTGTTTATTTGTTGTTATGGAAATTTCGTCAGTACAAAAATTAACTGCTGTTAAAATTTAGCGGCTTNTTACATACATATTATTTTAAAATGTTACAAATTCTAAGATAAAATACTAAGCTATTATATTATTTCTCCCCAGATGTCGGAAAAAAATTTTAAAAAATTATCAGNATCAGAAAACAGAAAATGGAAAGCCATTAGAGAAGAAAATGAAAAAAAATTATTTGGATCATTGAATAAATTTTTGGAGAAAATAGATGAAATGCCAGAAAAAAAGGAAAAGACTTCAGAGCTACTAGTTTCAGCAGAATGTAGCACAGATACGTCACTGATGGAGCAGGATGTGAATTTACCTTCTACTTCCATGTCAACCATAACTGATGAATTTCAAGAAAAACTGGAAGAGAAAAAATCAGAAGAATCCATTAAACCGAATGTACTTTCTGCCGATCCAGGTACGTGGCCATGTAATTTGAATATTAAACAACGGNACACTCTTGTTGAAAATTATCCACCTCAAGTAAGAAATTTTAACTTTCCAAAAGACAGTACAGGTAGGAAGTTTTCGGAAACTTATTANACACGAATTCTTCCAAATGGTGAAAAAATCACTAGATCCTGGTTACTTTATTCTACCTCAAAGGATTCTGTATTTTGTCTATATTGCAAACTCTTTGGGGATAGAAAAAATCAACTGAAGAATGAGAATGGATGCAAAGATTGGCAGCATTTATCACATATTCTTAGTAAACATGAAGAAAGTGAAATGCACATCAACAATAGTGTTAAGTATTCAAAATTGAAATCTGATTTGAAAAAATATATAACTATCGATGCTGCAGAACAGAGACTATATGAAAATGAGGAAAAATATTGGCGTGCTGTTGTTGAACGCATAATATACATGATTCAGTTTTTGGCAAGGCAATGTCTAGCGTTCAGAGGATCATCCAAGAAANTTTATGAGCGTGATAATGGGAATTTTCTTCATTTGGCAGAAACCGTAGCAAAATTTGATAATATTATGGCAGAACATTTATGCAGAATACAAACATTGACACCAAATAATATTACACATTATTTAGGGGATGAAATACAAAACCAAATAATTAAAATTATAGCAGATGCGATGAATGAAATCATTTTGAATAAGGTAAGAAATNCTAAATATTACTCAATTATTTTAGATACAACCCCTGATGTAAGCCATGTAGAACAGATAACAATTATTATTAGATTTGTGGATTTCCAAAATGCAAATATTTCTATATGTAAACATTTTGGGGGATTTTGTCCAGTAGATGACACAACTGGAGAAGGATTATATACATTTTTAGTAGACTACTTACAAAAACCGAATCTAGACAAAAATAATTTGAGAGGCCAGGGATACGACAATGCCACAAATATGCATGGAAAACATAACGGATTGCAACAGAAGATCCTCGAAAAATATTCTAGGGCATTCTATATTCCTTGTTCTGCGCATTCTTTAAACTTGGTAGTGAACGATGCAGCAAAAATCTCTTTTGACACCACTGAATTTTTTTATATNATCCAAGAACTATACAACTACTTTTCAGCTTCAACGAAACGTTGGAGTGTATTGAAAACACATTTACCAAACATAAATTTGAAACCATTGTCGAATACGCGATGGGAAAGTCGAGTTGATGCCATCATGCCACTAAAAACTCAATTAGAGGCAGTATGCAATGCTCTTTTAGAAATTNTTGATGATATTAATAAAGATATAGATTCTAGAATACAAGCCAAAGNTATACTTTCGAAAATATCCACTTTTAAATTTATTTGCTCAGTACTAATTTGGCATGATGTTTTAAGTAAAATTAATGTTATAAGCAAAATGATGCAGGAACCAAAATTCAATGTACATAATGCCCTTGAAAGCATCAAAGGGATTAAGAAATTCTTTGAAGAAAAACGAAGCGATGATGTTTATGAAAAATATATTATTGATGCTAAACAAATTGCAGAGTCTCTTGACATTGACGCCAACTTCTCCAATGTAGAAAATACTAGAAAAAGGAGAAAGAAGACTCATTTCGACTGCAAATCCCACGATCAGCCCATCTATGATCCAAAATTAAATTTCAAAATCAATTTCTTTTTTTCTATATTGGATACAACAATATCATCTTTAAAAGANAGGTTTACGATACCGAAAGAATATCATGGCCTTTTCAGTGTTCTAGATGACATGTGTAATTTGAAGAATTATAATGAATATGAACTACTGCAACTATGTAAAAATTTACAATTGAAATTGACTGATGAACATAGTAAGGAATGTGATATTGATGGGACATCAACGTACGAGGAACTAAAANCCTTAAGTTGTTTTCTAAAATCTAGTATNACTCCCGTTGAAGTATTGCAATATATTTGCAGCAATAATTTACCGGACACATTTCCGAATACTTTTGTAGCTCTCAGGATATATTTAACTTTGCCAATATCGGTCGCCAGTGGAGAACGTAGCTTCTCAAAATTAAAATTTATTAAAAACTACTTAAGGTCTACAATGTCACAAGAAAGACTGTCAAATCTAGCGACCATTTCTATAGAAAACGAGATATTGGAGCAAGTAAGCATTGAAACTGTCATAAAGAATTTTATTTCCATGAAAGCAAGACGTGTAACTATTTTATAACAAATTATTATATCCTAGTGACTTTGAGTTCCTAATTATTGTGATTAAAANTTCGAATTTTCTATTATTTTATGTTCAATAATTTGATTTGTATAATTTATACAGAATGACTTTATATTTGTTGTAATTGTTATTATTGAGAAATAAATAATATACTAACTTNAAAATTTTACTTGAGGAAGGGGCGCCTTTTTCTAATTCGCACAAAGGCGCCGTATGGGCTAGCAGCGGCCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Zaphod2 | ZNF558 | 1067 | 1095 | + | 0.05 | ATTTCAAGAAAAACTGGAAGAGAAAAAAT |
| Zaphod2 | Sox21b | 2831 | 2845 | + | -0.80 | AACAATATCATCTTT |
| Zaphod2 | THI2 | 2608 | 2622 | + | -2.21 | AGAAATTCTTTGAAG |
| Zaphod2 | BPC6 | 2802 | 2822 | + | -2.34 | ATCAATTTCTTTTTTTCTATA |
| Zaphod2 | BCL6B | 3319 | 3335 | - | -8.76 | TGCTTTCATGGAAATAA |
| Zaphod2 | AT4G12670 | 1574 | 1602 | - | -16.25 | CCAAAAACTGAATCATGTATATTATGCGT |
| Zaphod2 | BPC5 | 2796 | 2825 | - | -48.79 | CAATATAGAAAAAAAGAAATTGATTTTGAA |
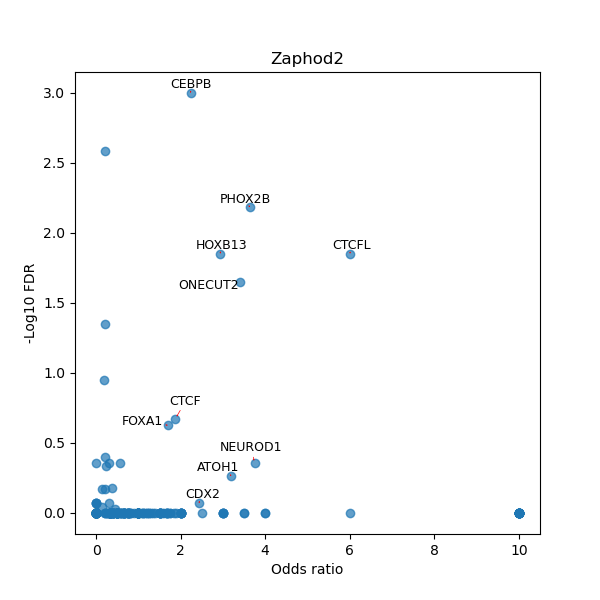
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.