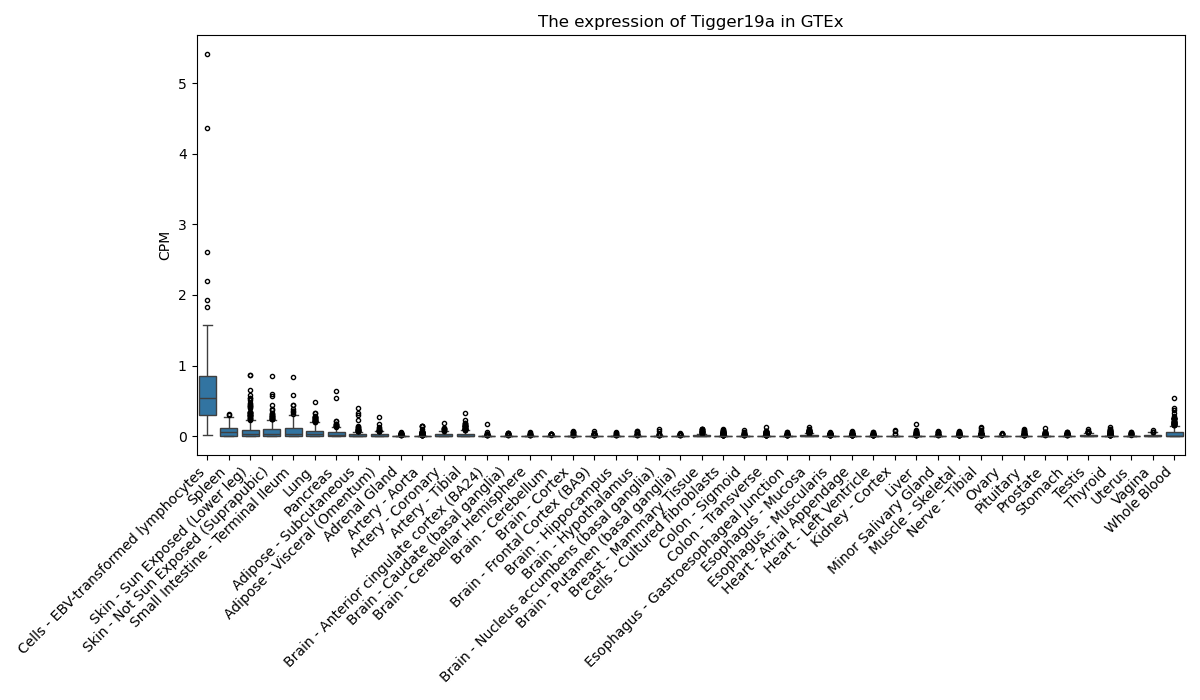
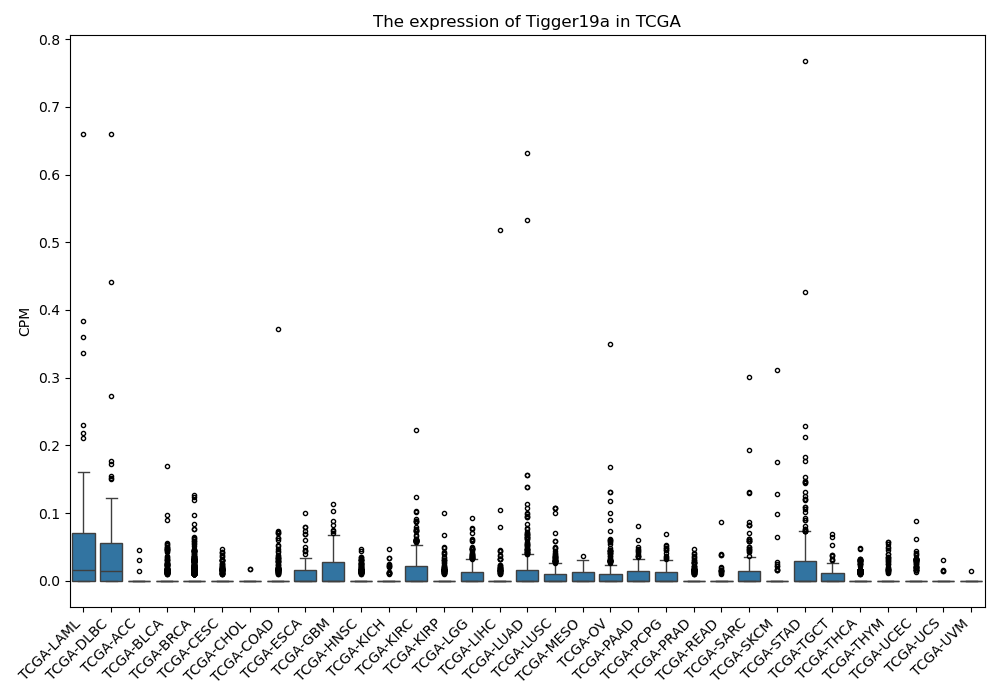
Tigger19a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000835 |
|---|---|
| TE superfamily | Tigger |
| TE class | DNA |
| Species | Theria_mammals |
| Length | 323 |
| Kimura value | 34.37 |
| Tau index | 0.9914 |
| Description | TcMar-Tigger DNA transposon, Tigger19a subfamily (non-autonomous) |
| Comment | Tigger19a is an older Tigger element, with "TA" TSDs. The termini are similar to autonomous SMAR15 Tigger element in the flatworm Schmidtea. There are about 2000 recognizable on average 35% substituted copies in the reconstructed laurasiatherian genome. |
| Sequence |
CAGTCAACTCTCGATTATCCGCGTGTGGATTATCCACTTTGTGGATTATCCGCGGCGAATTTTAAATCCCAGGCCGGCTTCCCCTCGCCACCCAGCATGCTTCCGGGCCGCTTCCTCCCGCCATCAGTTGGTGTGTGCTCAGGGAATGCAAACTAATTTTAATATTAGCCTAAGCAAAACATAATTAGGAAGGTAAATCTGCTGCGAAAAAATCACATAATGCATTCCAAAGCCAAATANAAATGATTTTTGGATTATCCGCTGTTTTGGATTATCCGCGCCACTGCTCCCCTCCATTAGCGCGGATAATCGAGAGTTGACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Tigger19a | WRKY42 | 2 | 9 | - | 12.57 | AGTTGACT |
| Tigger19a | WRKY42 | 315 | 322 | + | 12.57 | AGTTGACT |
| Tigger19a | AT1G19000 | 27 | 39 | - | 12.56 | AAGTGGATAATCC |
| Tigger19a | PK24580.1 | 72 | 78 | - | 12.56 | GCCGGCC |
| Tigger19a | POU5F1 | 146 | 152 | + | 12.44 | ATGCAAA |
| Tigger19a | Ets96B | 99 | 107 | - | 12.41 | CCCGGAAGC |
| Tigger19a | Zfp809 | 66 | 74 | + | 12.37 | ATCCCAGGC |
| Tigger19a | WRKY6 | 2 | 9 | - | 12.36 | AGTTGACT |
| Tigger19a | WRKY6 | 315 | 322 | + | 12.36 | AGTTGACT |
| Tigger19a | ERF6 | 72 | 78 | + | 12.31 | GGCCGGC |
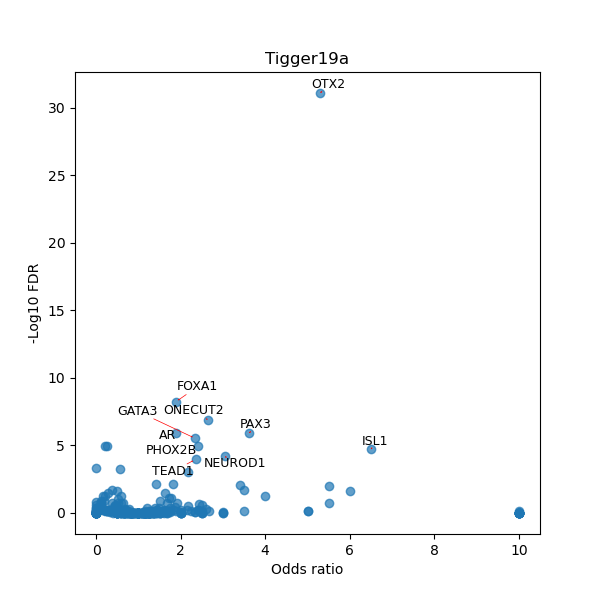
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.