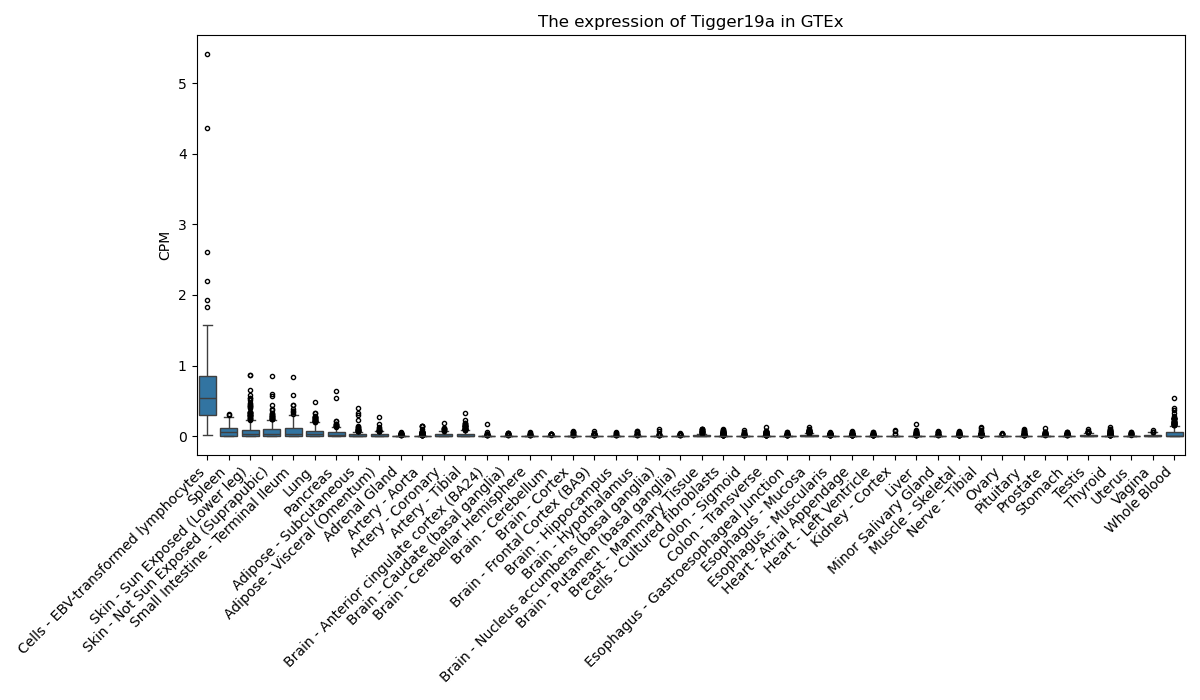
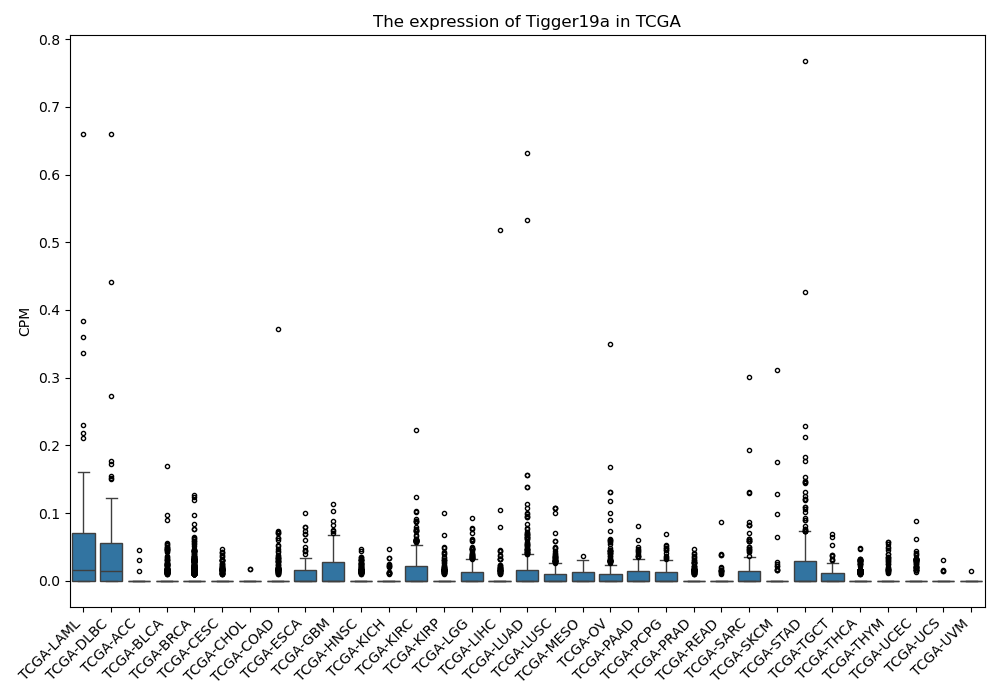
Tigger19a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000835 |
|---|---|
| TE superfamily | Tigger |
| TE class | DNA |
| Species | Theria_mammals |
| Length | 323 |
| Kimura value | 34.37 |
| Tau index | 0.9914 |
| Description | TcMar-Tigger DNA transposon, Tigger19a subfamily (non-autonomous) |
| Comment | Tigger19a is an older Tigger element, with "TA" TSDs. The termini are similar to autonomous SMAR15 Tigger element in the flatworm Schmidtea. There are about 2000 recognizable on average 35% substituted copies in the reconstructed laurasiatherian genome. |
| Sequence |
CAGTCAACTCTCGATTATCCGCGTGTGGATTATCCACTTTGTGGATTATCCGCGGCGAATTTTAAATCCCAGGCCGGCTTCCCCTCGCCACCCAGCATGCTTCCGGGCCGCTTCCTCCCGCCATCAGTTGGTGTGTGCTCAGGGAATGCAAACTAATTTTAATATTAGCCTAAGCAAAACATAATTAGGAAGGTAAATCTGCTGCGAAAAAATCACATAATGCATTCCAAAGCCAAATANAAATGATTTTTGGATTATCCGCTGTTTTGGATTATCCGCGCCACTGCTCCCCTCCATTAGCGCGGATAATCGAGAGTTGACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Tigger19a | DPRX | 252 | 260 | - | 13.02 | GGATAATCC |
| Tigger19a | DPRX | 269 | 277 | - | 13.02 | GGATAATCC |
| Tigger19a | DPRX | 27 | 35 | - | 13.02 | GGATAATCC |
| Tigger19a | DPRX | 43 | 51 | - | 13.02 | GGATAATCC |
| Tigger19a | TEAD3 | 222 | 229 | + | 12.98 | GCATTCCA |
| Tigger19a | rib | 203 | 211 | + | 12.74 | TGCGAAAAA |
| Tigger19a | MAZ | 289 | 296 | + | 12.71 | CCCCTCCA |
| Tigger19a | Eip74EF | 100 | 106 | - | 12.66 | CCGGAAG |
| Tigger19a | bowl | 99 | 106 | + | 12.66 | GCTTCCGG |
| Tigger19a | Isl1 | 294 | 300 | + | 12.65 | CCATTAG |
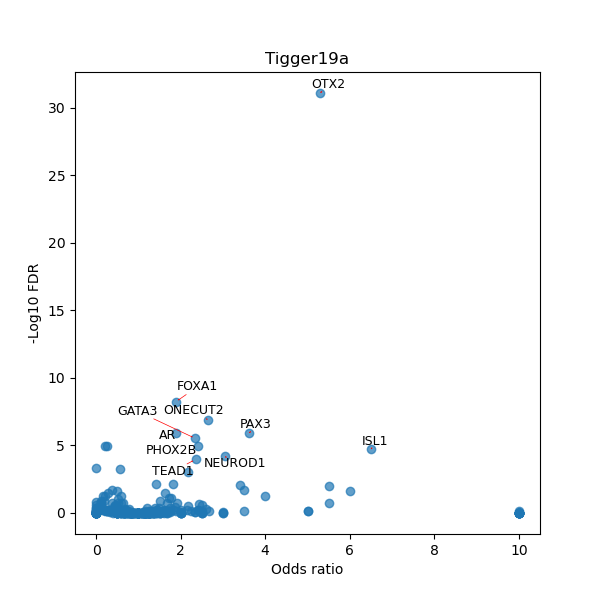
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.