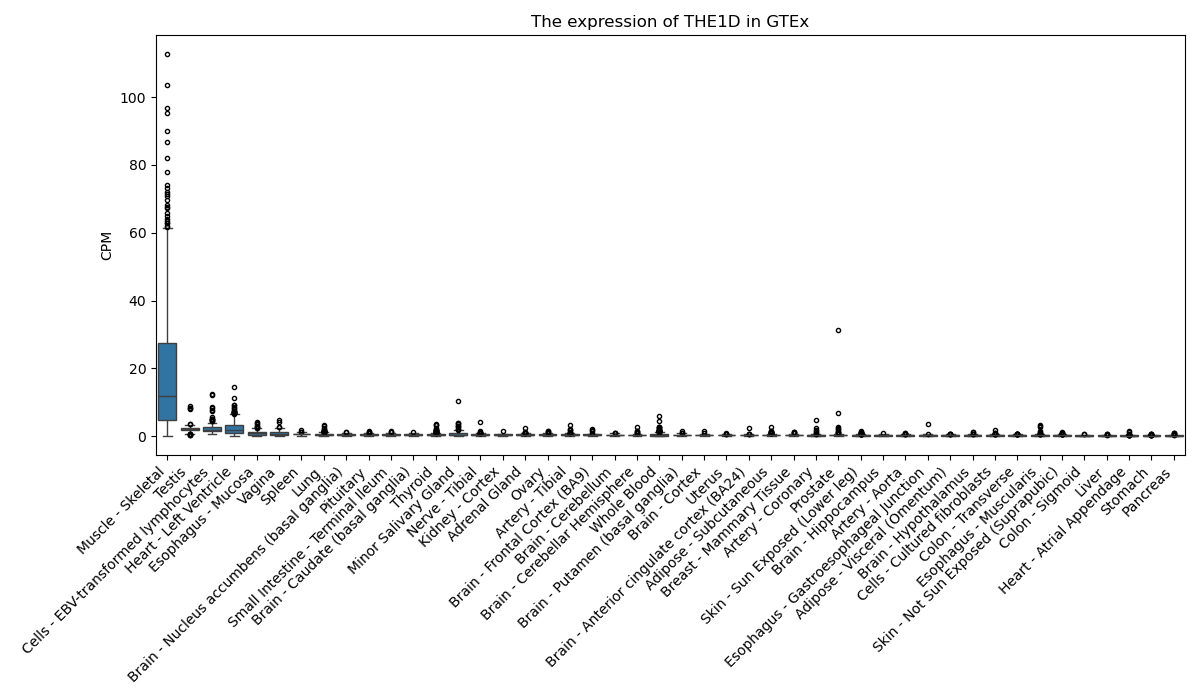
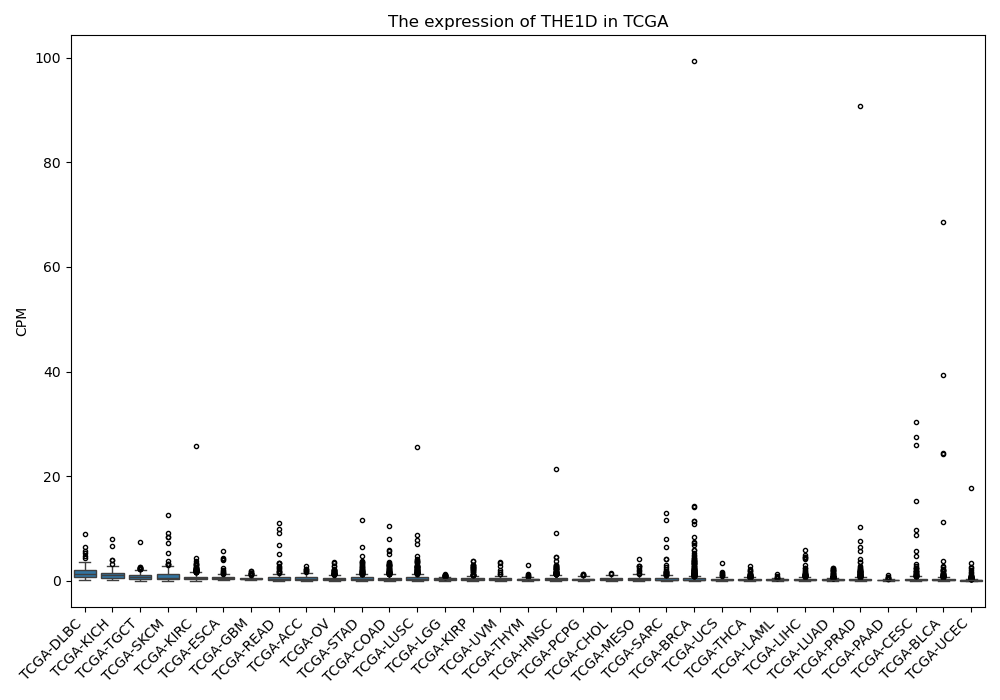
THE1D
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000820 |
|---|---|
| TE superfamily | MaLR |
| TE class | LTR |
| Species | Haplorrhini |
| Length | 381 |
| Kimura value | 10.85 |
| Tau index | 0.9642 |
| Description | THE1D Long Terminal Repeat for ERVL-MaLR endogenous retrovirus |
| Comment | THE1D is 80% similar to THE1A, 84% to THE1B, and 89% to THE1C. |
| Sequence |
TGATATGGTTTGGCTGTGTCCCCACCCAAANTCTCATCTCGAATTGTAATCCCCATAATCCCCACGTGTCGAGGGAGGGACCTGGTGGAGGTGATTGGATCATGGGGGCGGTTTCCCCCATGCTGTTCTCGTGATAGTGAGTGAGTTCTCACGAGATCTGATGGTTTTATAAGTGTCTGGCAGNTTTCCCCTGCNCTCACNCTTCTCTCTCCTGCCGCCNTGTGAAGAAGGTGCTTGCTTCCCCTTCGCCTTCCGCCATGATTGTAAGTTTCCTGAGGCCTCCCCAGCCATGCGGAACTGTGAGTCAATTAAACCTCTTTCCTTTATAAATTACCCAGTCTCGGGTAGTTCTTTATAGCAGTGTGAAAACGGACTAATACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| THE1D | EmBP-1 | 62 | 69 | - | 14.49 | ACACGTGG |
| THE1D | ABI5 | 63 | 70 | - | 14.41 | GACACGTG |
| THE1D | Mnt | 62 | 68 | + | 14.31 | CCACGTG |
| THE1D | LRL2 | 54 | 68 | + | 14.29 | CATAATCCCCACGTG |
| THE1D | ZKSCAN5 | 87 | 95 | + | 14.28 | GGAGGTGAT |
| THE1D | ref-1 | 62 | 69 | - | 14.08 | ACACGTGG |
| THE1D | KLF5 | 19 | 28 | + | 14.04 | TCCCCACCCA |
| THE1D | dm | 61 | 68 | + | 14.03 | CCCACGTG |
| THE1D | Max | 61 | 68 | + | 14.03 | CCCACGTG |
| THE1D | btd | 104 | 112 | + | 14.02 | GGGGGCGGT |
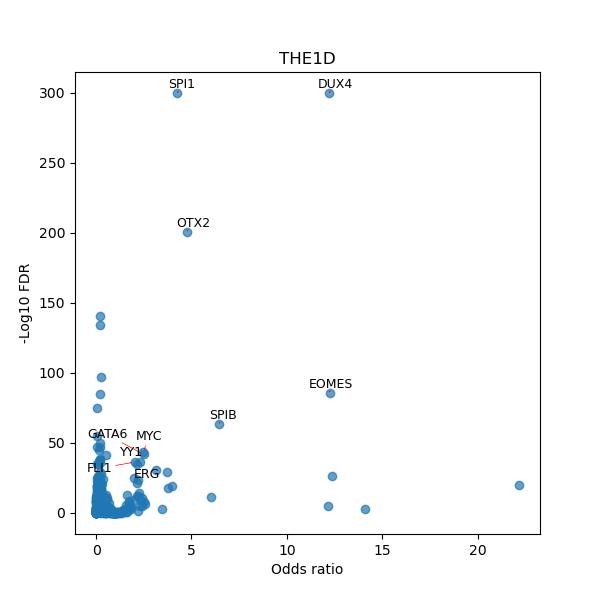
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.