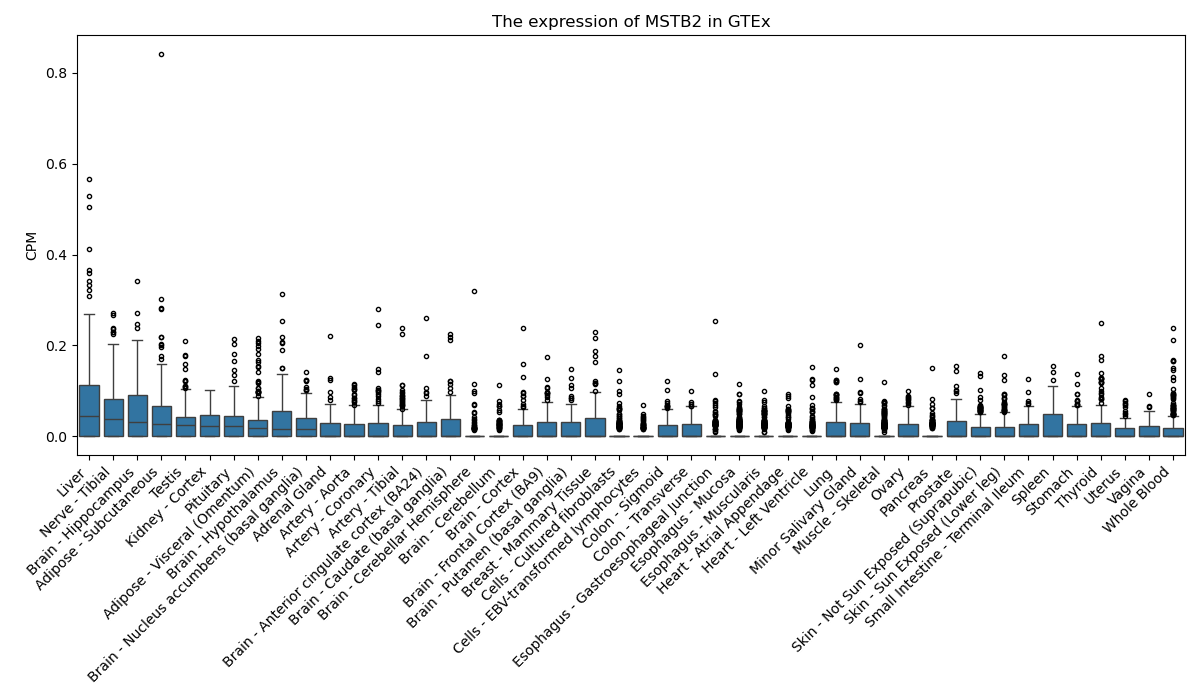
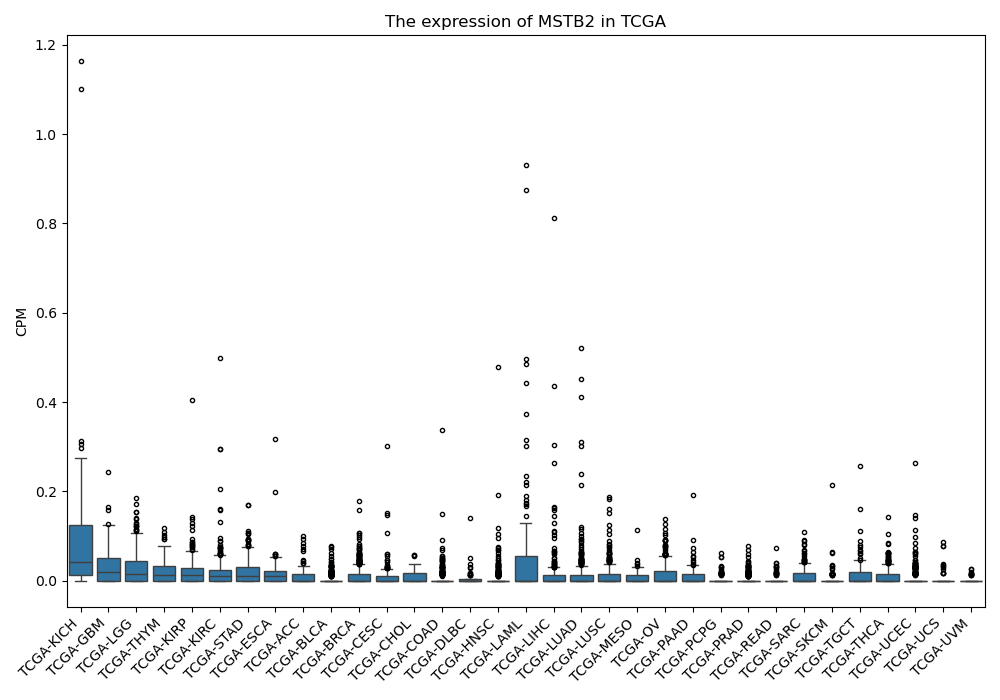
MSTB2
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001134 |
|---|---|
| TE superfamily | MaLR |
| TE class | LTR |
| Species | Primates |
| Length | 457 |
| Kimura value | 16.06 |
| Tau index | 0.8954 |
| Description | MSTB2 Long Terminal Repeat for ERVL-MaLR retrotransposon |
| Comment | Note that a full-length ERV-MaLR (Mammalian apparent LTR retrotransposon) element is derived from an ERV (Endogenous Retrovirus), but only contains the Gag gene (that is, no Pol or Env). |
| Sequence |
TGCTATGGTTTGGATATGGTTTGTTTGTCCCCACCAAAACTCATGTTGAAATTTGATCCCCAATGTGGCGGTGTTGGGAGGTGGGGCCTAGTGGGAGGTGTTTGGGTCATGGGGGCGGATCCCTCATGAATGGCTTGGTGCCGTTCTCGCGGTAGTGAGTGAGTGAGTTCTCGCTCTCGCGAGACTGGATTAGTTCTCGCGGGAATGGATTAGTTCCCGCGAGAGTGGGTTGTTATAAAGCCAGGACGCCCCTCGGGTTTTGCCTCTTCGCACGTGTCCGCTTCCCCTTTGACCTTCTCCGCCATGTTNTGACGCAGCACGAAAGCCCTCACCAGAAGCCAGGGCCATGCCCTTGAACTTCCCAGCCTGCAGAACCGTGAGCTAAATAAACCTCTTTTCTTTATAAATTACCCAGTCTCAGGTATTCTGTTATAGCAACACAAAACGGACTAAGACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
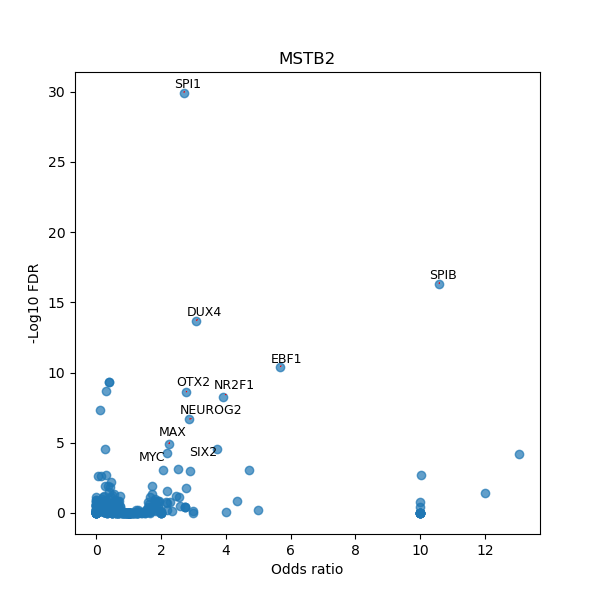
| MSTB2 | HEY2 | 270 | 278 | - | 15.33 | GACACGTGC |
| MSTB2 | ZBTB33 | 195 | 204 | - | 15.17 | TCCCGCGAGA |
| MSTB2 | ZBTB33 | 215 | 224 | + | 15.17 | TCCCGCGAGA |
| MSTB2 | PATZ1 | 77 | 87 | + | 15.04 | GGAGGTGGGGC |
| MSTB2 | FEZF2 | 361 | 368 | + | 15.03 | CCCAGCCT |
| MSTB2 | PLAG1 | 83 | 96 | + | 15.01 | GGGGCCTAGTGGGA |
| MSTB2 | ref-1 | 270 | 277 | - | 14.90 | ACACGTGC |
| MSTB2 | BZR2 | 270 | 279 | + | 14.83 | GCACGTGTCC |
| MSTB2 | Bteb2 | 109 | 121 | - | 14.79 | GATCCGCCCCCAT |
| MSTB2 | Spi1 | 278 | 290 | - | 14.77 | AAAGGGGAAGCGG |
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.