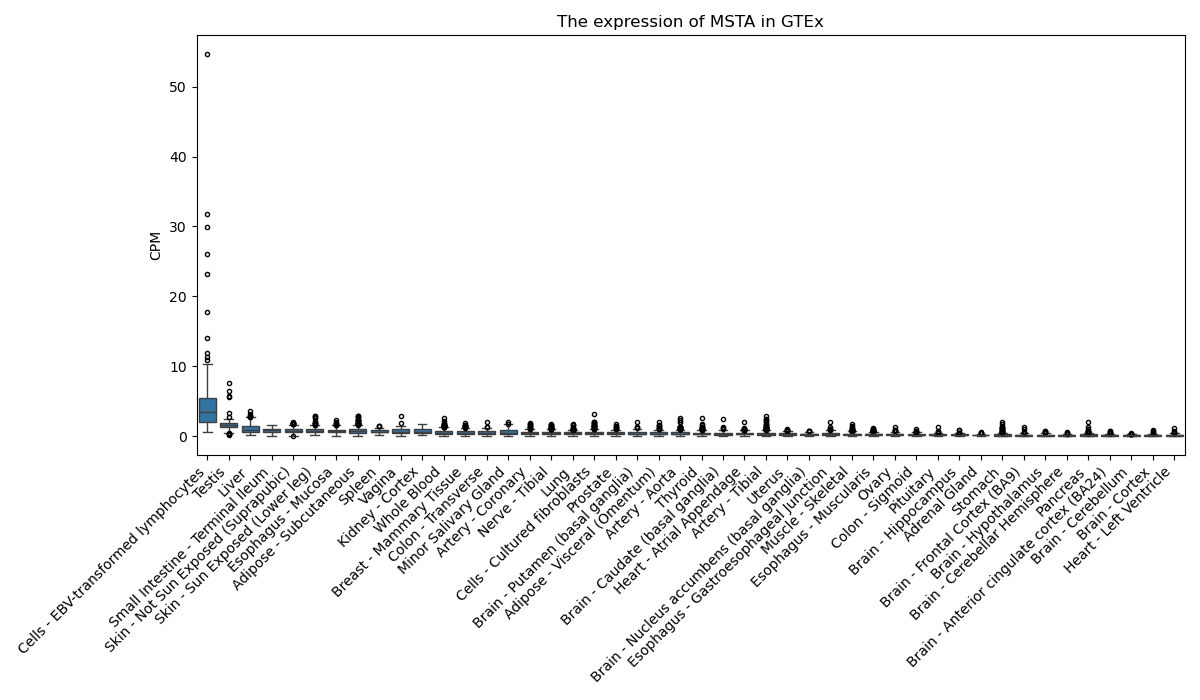
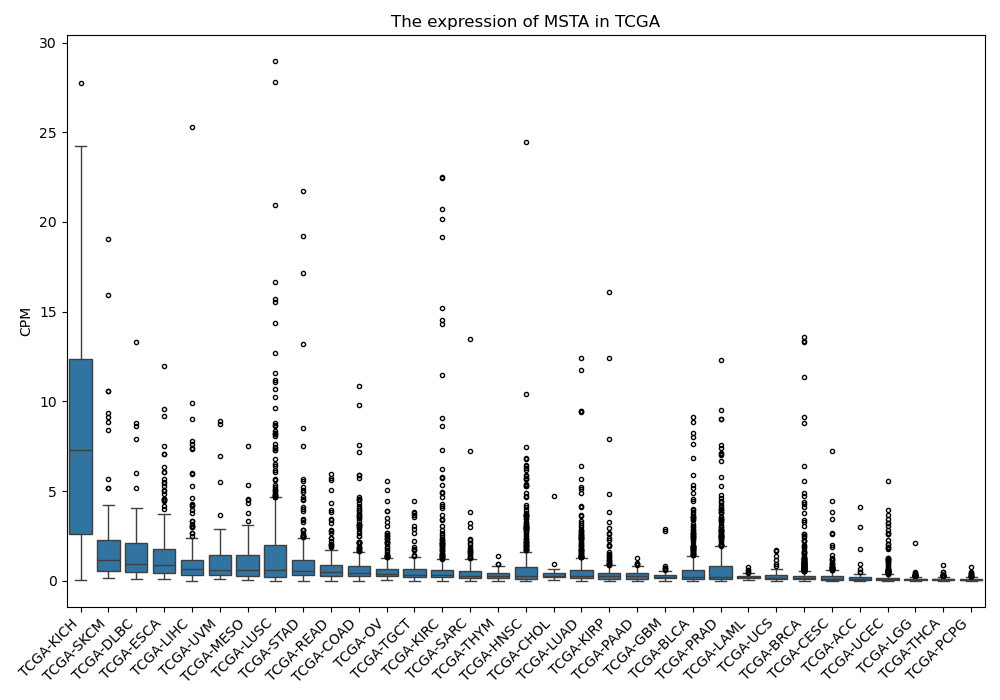
MSTA
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001041 |
|---|---|
| TE superfamily | MaLR |
| TE class | LTR |
| Species | Haplorrhini |
| Length | 428 |
| Kimura value | 12.64 |
| Tau index | 0.8869 |
| Description | MSTA Long Terminal Repeat for ERVL-MaLR retrotransposon |
| Comment | MSTA has 5 bp TSDs. This represents an intermediate subfamily between THE1C and MSTB. Note that a full-length ERV-MaLR (Mammalian apparent LTR retrotransposon) element is derived from an ERV (Endogenous Retrovirus), but only contains the Gag gene (that is, no Pol or Env). |
| Sequence |
TGATATGGTTTGGATCTGTGTCCCCGCCCAAATCTCATGTCGAATTGTAATCCCCAGTGTTGGAGGNGGGGCCTGGTGGGAGGTGATTGGATCATGGGGGCGGATTTCTCATGANCGGTTTAGCACCATCCCCTTGGTGCTGTTCTCGTGATAGTGAGTGAGTTCTCACGAGATCTGGTTGTTTAAAAGTGTGTGGCACCTCCCCCCTCGCTCTCTCTTNCTCCTGCTCCGGCCATGTGACGTGCCTGCTTCCCCTTCGCCTTCCGCCATGATTGTAAGTTTCCTGAGGCCTCCCCAGAAGCCGANNAGATGCCANCGCCATGCTTCCTGTACAGCCTGCGGAACCGTGAGCCAATTAAACCTCTTTTCTTTATAAATTACCCAGTCTCAGGTATTTCTTTATAGCAGTGCGAGAACGGACTAATACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| MSTA | ZNF454 | 337 | 353 | - | 15.28 | GGCTCACGGTTCCGCAG |
| MSTA | Dux | 84 | 94 | - | 15.02 | TGATCCAATCA |
| MSTA | KLF10 | 22 | 30 | - | 14.62 | TGGGCGGGG |
| MSTA | MZF1 | 49 | 56 | + | 14.60 | AATCCCCA |
| MSTA | ZNF701 | 201 | 217 | - | 14.51 | GAGAGAGCGAGGGGGGA |
| MSTA | KLF5 | 21 | 30 | + | 14.37 | TCCCCGCCCA |
| MSTA | GATA1 | 171 | 180 | + | 14.30 | AGATCTGGTT |
| MSTA | ZKSCAN5 | 79 | 87 | + | 14.28 | GGAGGTGAT |
| MSTA | ZNF701 | 199 | 215 | - | 14.22 | GAGAGCGAGGGGGGAGG |
| MSTA | Fer2 | 233 | 242 | + | 14.21 | CCATGTGACG |
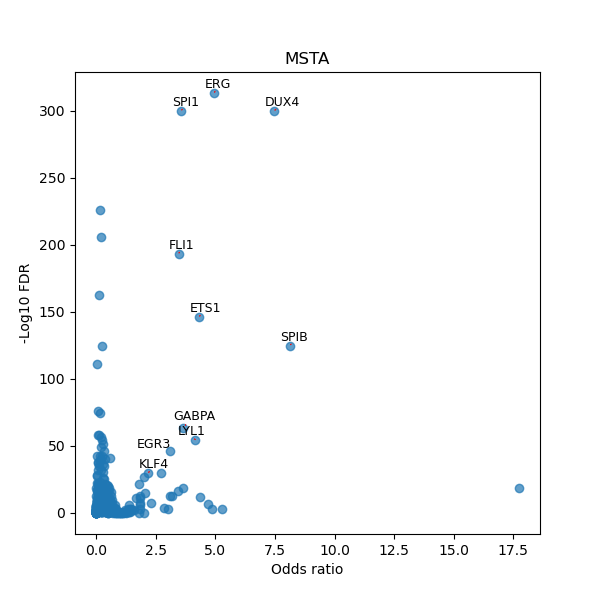
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.