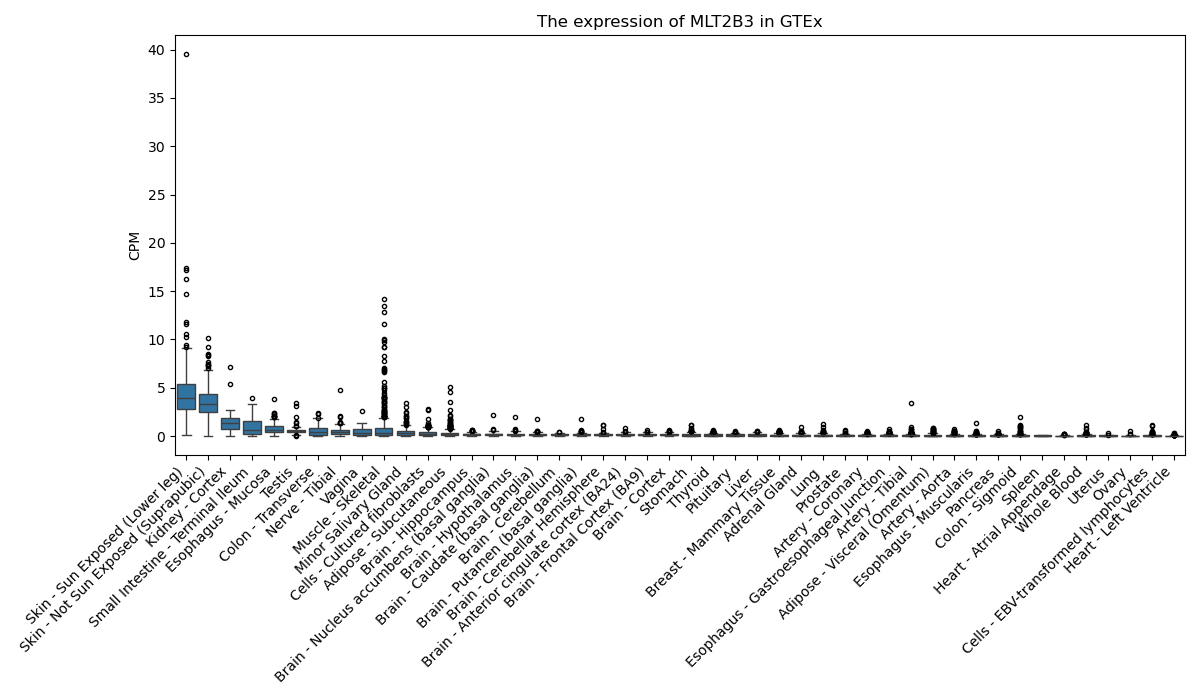
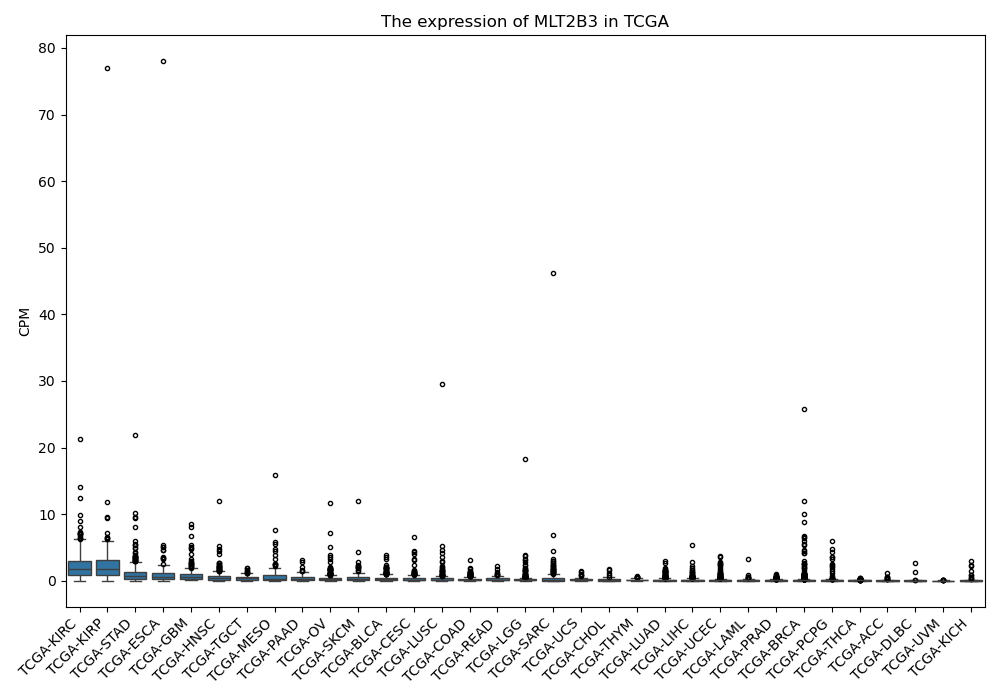
MLT2B3
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001031 |
|---|---|
| TE superfamily | ERV3 |
| TE class | LTR |
| Species | Primates |
| Length | 554 |
| Kimura value | 14.74 |
| Tau index | 0.9340 |
| Description | MLT2B3 Long Terminal Repeat for ERVL endogenous retrovirus |
| Comment | MLT2B3 is an ERVL LTR. However, a related LTR was "absorbed" by a mobile Ricksha DNA transposon; explaining this cross-Type homology. MLT2B3 contains a 50bp insertion compared to the otherwise closely related MLT2B2 variant. |
| Sequence |
TGTGATGGTTAATATTAGGTGTCAACTTGATTGGATTGAAGGATGCCNAGATAGCTGGTAAAGTATTGTTTCTGGGTGTGTCTGTGAGGGTGTTGCCAGAGGAGATTAACATTTGAGTCAGTGGACTGGGAGAGGAAGACCCACCCTCANCGATGTGGGTGGGCACCATCCAATCGGCTGCCGGCNCGGCTAGAACAAAAAGGCAGAGGAAGGGCGAATTTGCTCTCTCTCCTGGAGCTGGGACACCCTTCTCCTCCTGCCCTCGGACATCAGANCTCCAGGTTCTTCGGCCTTTGGACTCTNGGACTTACACCAGCGGTTTNCCGGGCTCTCGGGCCTTCGGCCACAGACTGAAGGCTGCACCGTCGGCTTCCCTGCTTTTGAGGCTTTNGGACTCGGACTGAGCCACGCTACCGGCTTCCCTGGCTCCCCAGCTTGCAGACGGCCTATCGTGGGACTTCNCGGCCTTCGTGATCGTGTGAGCCAATTCCCCTAATAAATCCCCTTTATCTATCCTATTAGTTCTGTCCCTCTGGAGAACCCTGACTAATACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| MLT2B3 | KLF9 | 72 | 82 | - | 14.09 | GACACACCCAG |
| MLT2B3 | TBX3 | 17 | 25 | + | 13.87 | AGGTGTCAA |
| MLT2B3 | klu | 152 | 162 | - | 13.79 | CCACCCACATC |
| MLT2B3 | fos-1 | 114 | 120 | - | 13.76 | TGACTCA |
| MLT2B3 | ERF6 | 179 | 185 | + | 13.72 | TGCCGGC |
| MLT2B3 | DUX4 | 28 | 38 | - | 13.70 | CAATCCAATCA |
| MLT2B3 | PK24580.1 | 179 | 185 | - | 13.69 | GCCGGCA |
| MLT2B3 | odd-2 | 410 | 417 | + | 13.67 | GCTACCGG |
| MLT2B3 | Klf6-7-like | 67 | 86 | - | 13.58 | CACAGACACACCCAGAAACA |
| MLT2B3 | MYB99 | 14 | 21 | - | 13.30 | CACCTAAT |
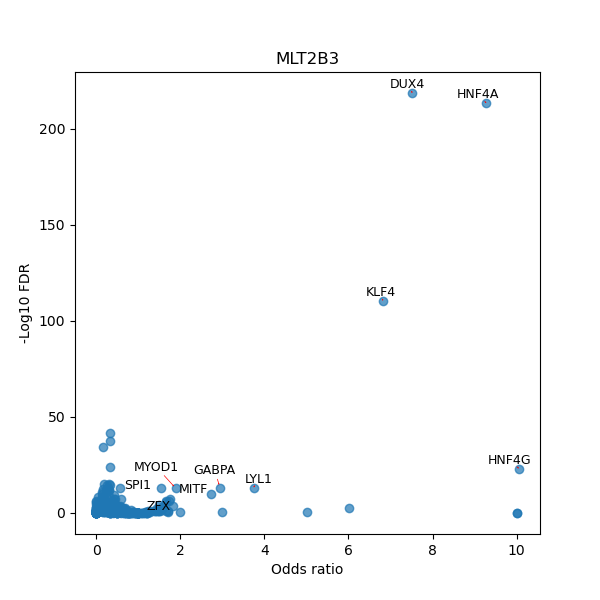
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.