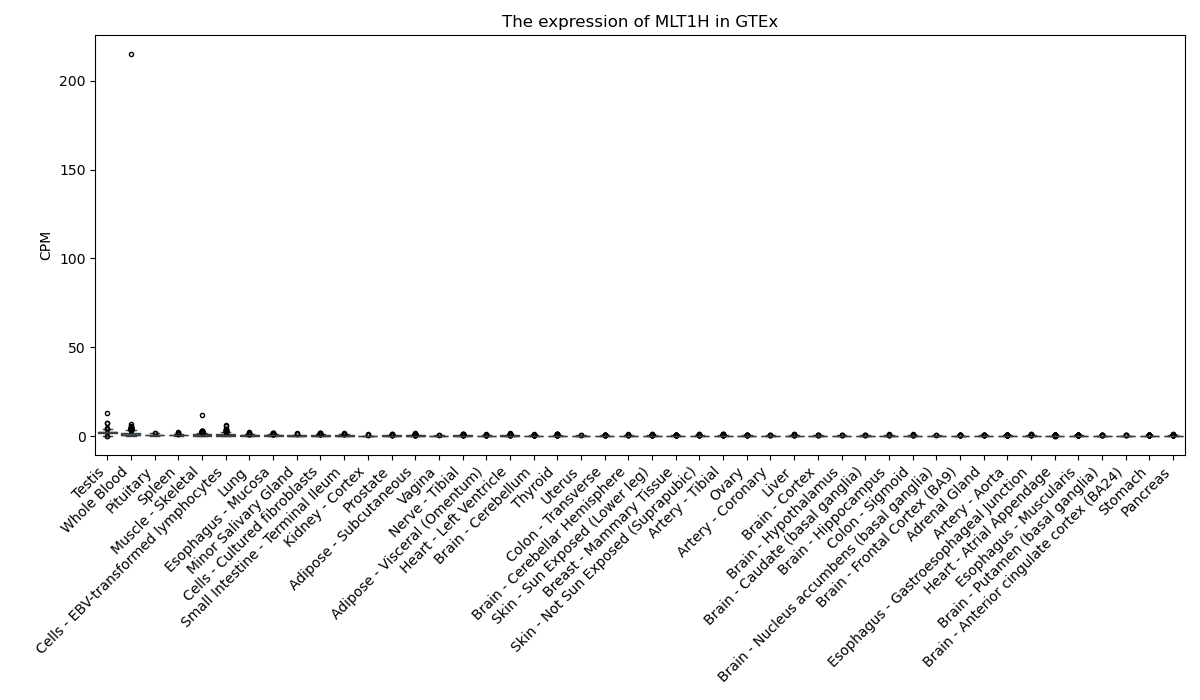
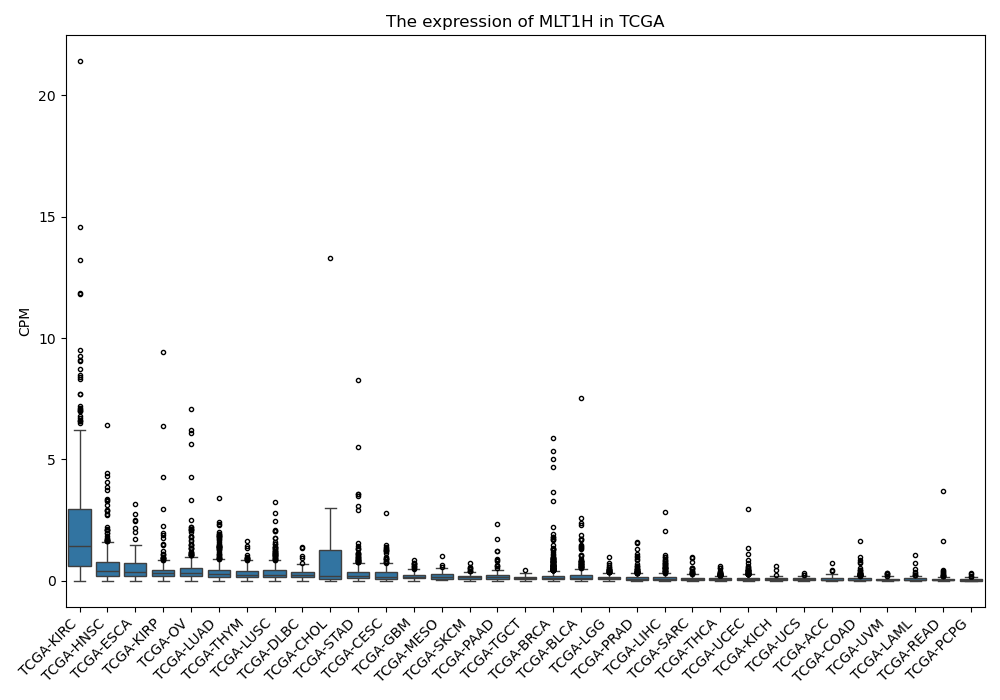
MLT1H
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001014 |
|---|---|
| TE superfamily | MaLR |
| TE class | LTR |
| Species | Eutheria |
| Length | 549 |
| Kimura value | 27.41 |
| Tau index | 0.8986 |
| Description | MLT1H Long Terminal Repeat for ERVL-MaLR retrotransposon |
| Comment | MLT1H has 5 bp TSDs. Note that a full-length ERV-MaLR (Mammalian apparent LTR retrotransposon) element is derived from an ERV (Endogenous Retrovirus), but only contains the Gag gene (that is, no Pol or Env). |
| Sequence |
TGTGGTAGATTGATTGCAAAAATGGCCNCAATTCTTCACCCCTCCCTGTATCCACGCCCTTTGCAATGTGACTTTGCAGCTCCTCCCATCAAGAGGTGGAGTCTATTTCCCCACCCCTTGAATCTGGGCTGGCCTTGTGACTTGCTTTGGCCAATAGAATGCGGCGGAAGTGACGNTGTGCCAGTTCCGAGCCTAGGCCTCAAGAGGCCTTGCACGCTTCCGCTCTCTCTCTTGGAACCCTGCCACCGCCATGTGAACAAGCCCGGGCTAGCCTGCTGGAGGATGAGAGACCACGTGGAGCAGAGCCGAGCCGTCCCAGCTGAGGCCATCCTAGACCAGCCAGCCCCCAGCCAAGATCAGCAGAGCCGCCTACCCGACCCGCAGCTGACCGCAGACGCATGAGTGAGCCCAGCCGAGACCAGAAGAACCGCCCAGCTGAGCCCAGCCCAAATTGCCGACCCACAGAATCGTGAGCTAAAATAAATGGTTGTTGTTTTAAGCCACTAAGTTTTGGGGTGGTTTGTTACGCAGCAATAGCTAACTGATACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| MLT1H | MYCN | 291 | 298 | - | 15.35 | CCACGTGG |
| MLT1H | MAX::MYC | 290 | 299 | + | 15.29 | ACCACGTGGA |
| MLT1H | KLF10 | 109 | 117 | - | 15.20 | GGGGTGGGG |
| MLT1H | dm | 290 | 297 | + | 15.13 | ACCACGTG |
| MLT1H | Max | 290 | 297 | + | 15.13 | ACCACGTG |
| MLT1H | Tfcp2l1 | 409 | 422 | + | 15.10 | CCAGCCGAGACCAG |
| MLT1H | ETV7 | 164 | 172 | + | 15.00 | GCGGAAGTG |
| MLT1H | KLF5 | 108 | 117 | + | 14.84 | TCCCCACCCC |
| MLT1H | DREB2A | 244 | 252 | - | 14.82 | ATGGCGGTG |
| MLT1H | Trl | 223 | 231 | - | 14.81 | GAGAGAGAG |
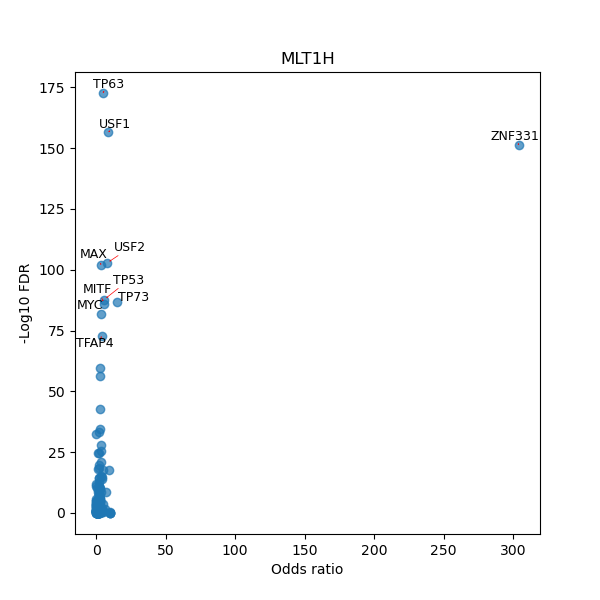
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.