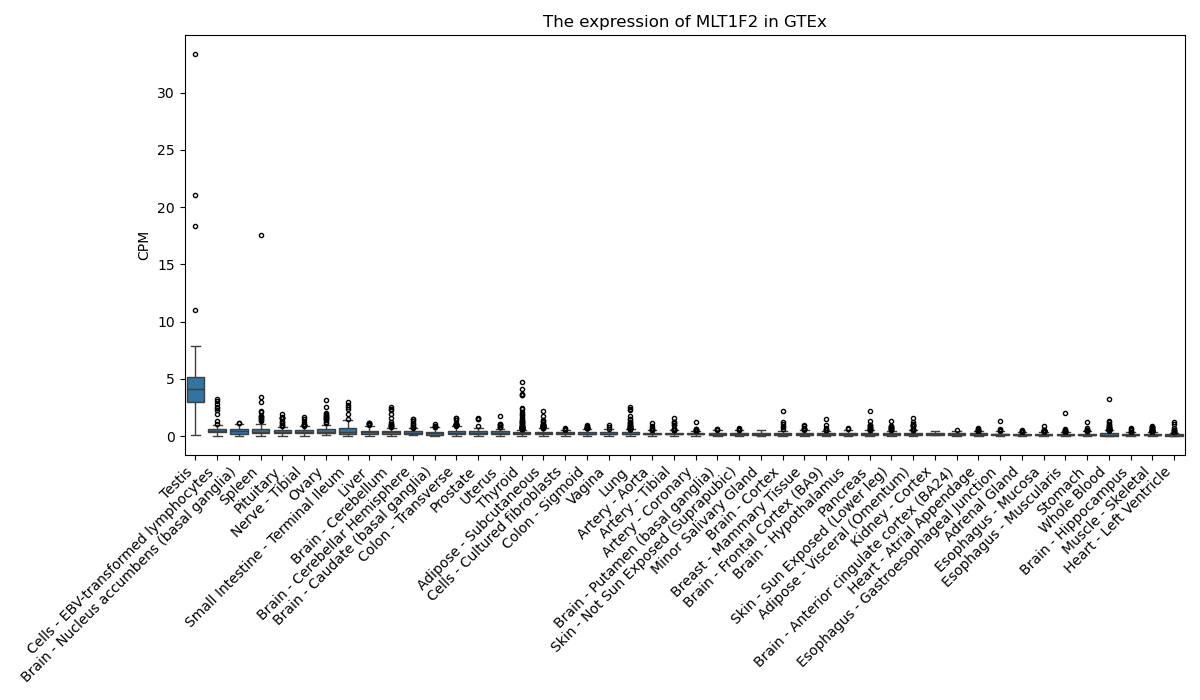
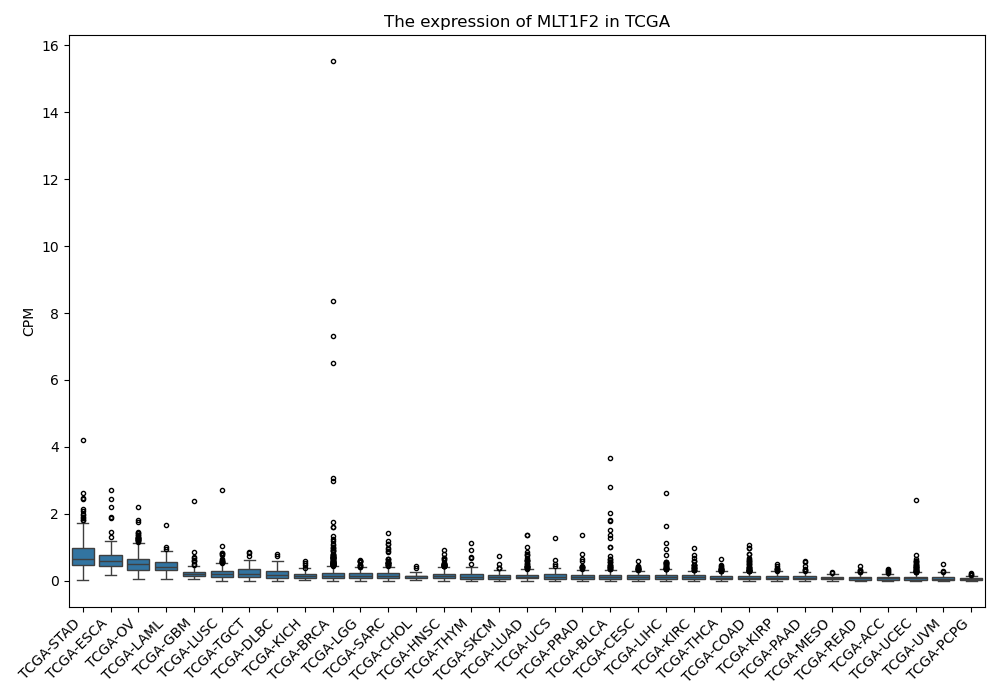
MLT1F2
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001010 |
|---|---|
| TE superfamily | MaLR |
| TE class | LTR |
| Species | Eutheria |
| Length | 560 |
| Kimura value | 23.89 |
| Tau index | 0.9445 |
| Description | MLT1F2 Long Terminal Repeat for ERVL-MaLR retrotransposon |
| Comment | MLT1F2 has 5 bp TSDs. Note that a full-length ERV-MaLR (Mammalian apparent LTR retrotransposon) element is derived from an ERV (Endogenous Retrovirus), but only contains the Gag gene (that is, no Pol or Env). |
| Sequence |
TGTGGTAGGCAGCCTCTAAGATGGCCCCCAATGATCCCCGCCTCCTGGTATTCACGCCCTTGTGTAATCCCCTCCCCTTGAGTGTGGGCTGGACCTAGTGACTCGCTTCTAACGAATAGAATACGGCAAAAGTGATGGGATGTCACTTCCGAGATTAGGTTACAAAAAGACTGTGGCTTCCGTCTTGCGCGCCCTCTCTTGCTCTCTCGCTTGCTCGCTCTGANGGAAGCNAGCTGCCATGTTGTGAGCTGCCCTATGGAGAGGCCCACGTGGCAAGGAACTGAGGGCGGCCTCCGGCCAACAGCCAGCGAGGAACTGAGNCCTGCCAACAACCACGTGAGTGAGCTTGGAAGCGGATCCTCCCCCAGTCGAGCCTTNAGATGACCGCAGCCCCGGCCGACACCTTGATTGCAGCCTCGTGAGAGACCCTGAGCCAGAGGCACCCAGCTAAGCCGCGCCCGGATTCCTGACCCACAGAAACTGTGAGATAATAAATGTNTGTTGTTTTAAGCCGCTAAGTTTTGGGGTAATTTGTTACGCAGCAATAGATAACTAATACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| MLT1F2 | BZIP16 | 266 | 275 | + | 17.23 | CCACGTGGCA |
| MLT1F2 | Clamp | 203 | 216 | - | 17.12 | GAGCAAGCGAGAGA |
| MLT1F2 | ABF1 | 266 | 275 | + | 16.72 | CCACGTGGCA |
| MLT1F2 | BHLH72 | 266 | 274 | - | 16.71 | GCCACGTGG |
| MLT1F2 | ZNF148 | 69 | 78 | + | 16.10 | CCCCTCCCCT |
| MLT1F2 | MAZ | 69 | 76 | + | 16.08 | CCCCTCCC |
| MLT1F2 | SP1 | 69 | 77 | - | 16.06 | GGGGAGGGG |
| MLT1F2 | BZIP68 | 266 | 275 | - | 15.97 | TGCCACGTGG |
| MLT1F2 | SP4 | 69 | 77 | - | 15.85 | GGGGAGGGG |
| MLT1F2 | KLF3 | 451 | 460 | + | 15.53 | AGCCGCGCCC |
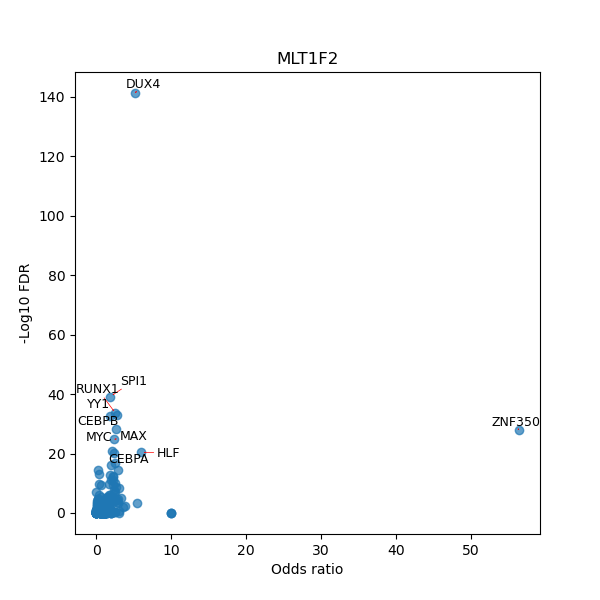
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.